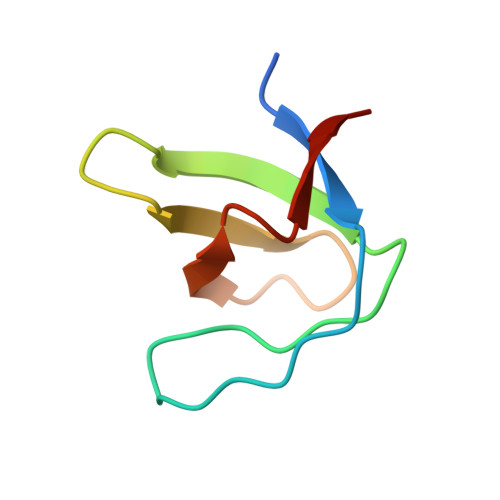
Crystallization of and selenomethionine phasing strategy for a SETMAR-DNA complex.
Chen, Q., Georgiadis, M.(2016) Acta Crystallogr F Struct Biol Commun 72: 713-719
- PubMed: 27599863
- DOI: https://doi.org/10.1107/S2053230X16012723
- Primary Citation of Related Structures:
4EIK - PubMed Abstract:
Transposable elements have played a critical role in the creation of new genes in all higher eukaryotes, including humans. Although the chimeric fusion protein SETMAR is no longer active as a transposase, it contains both the DNA-binding domain (DBD) and catalytic domain of the Hsmar1 transposase. The amino-acid sequence of the DBD has been virtually unchanged in 50 million years and, as a consequence, SETMAR retains its sequence-specific binding to the ancestral Hsmar1 terminal inverted repeat (TIR) sequence. Thus, the DNA-binding activity of SETMAR is likely to have an important biological function. To determine the structural basis for the recognition of TIR DNA by SETMAR, the design of TIR-containing oligonucleotides and SETMAR DBD variants, crystallization of DBD-DNA complexes, phasing strategies and initial phasing experiments are reported here. An unexpected finding was that oligonucleotides containing two BrdUs in place of thymidines produced better quality crystals in complex with SETMAR than their natural counterparts.
Organizational Affiliation:
Biochemistry and Molecular Biology, Indiana University School of Medicine, 635 Barnhill Drive, Indianapolis, IN 46202, USA.