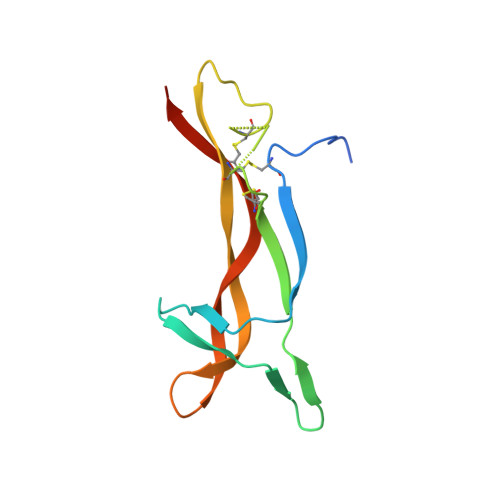
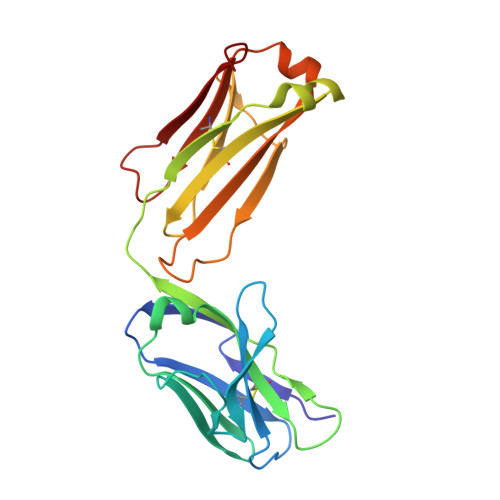
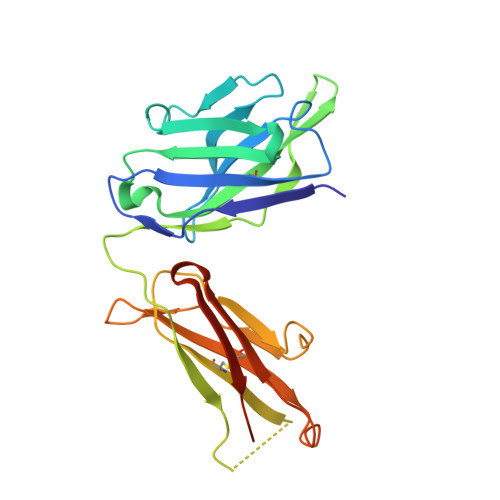
Generation of a high-fidelity antibody against nerve growth factor using library scanning mutagenesis and validation with structures of the initial and optimized Fab-antigen complexes.
La Porte, S.L., Eigenbrot, C., Ultsch, M., Ho, W.H., Foletti, D., Forgie, A., Lindquist, K.C., Shelton, D.L., Pons, J.(null) MAbs 6: 1059-1068
- PubMed: 24830649
- DOI: https://doi.org/10.4161/mabs.28677
- Primary Citation of Related Structures:
4EDW, 4EDX - PubMed Abstract:
Nerve growth factor (NGF) is indispensable during normal embryonic development and critical for the amplification of pain signals in adults. Intervention in NGF signaling holds promise for the alleviation of pain resulting from human diseases such as osteoarthritis, cancer and chronic lower back disorders. We developed a fast, high-fidelity method to convert a hybridoma-derived NGF-targeted mouse antibody into a clinical candidate. This method, termed Library Scanning Mutagenesis (LSM), resulted in the ultra-high affinity antibody tanezumab, a first-in-class anti-hyperalgesic specific for an NGF epitope. Functional and structural comparisons between tanezumab and the mouse 911 precursor antibody using neurotrophin-specific cell survival assays and X-ray crystal structures of both Fab-antigen complexes illustrated high fidelity retention of the NGF epitope. These results suggest the potential for wide applicability of the LSM method for optimization of well-characterized antibodies during humanization.