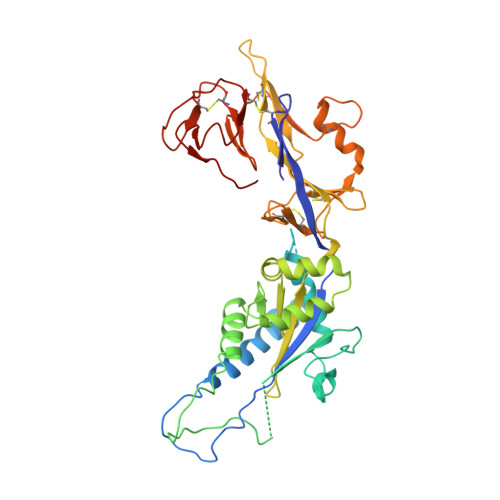
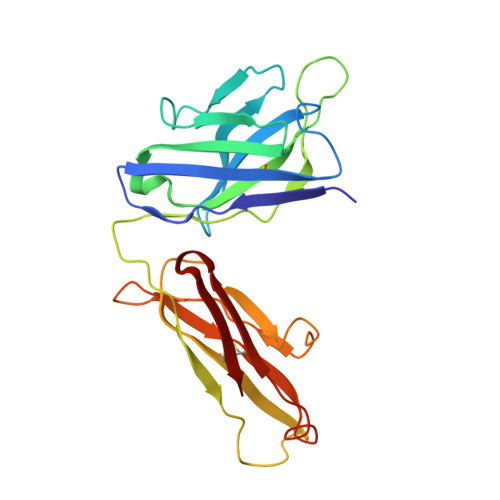
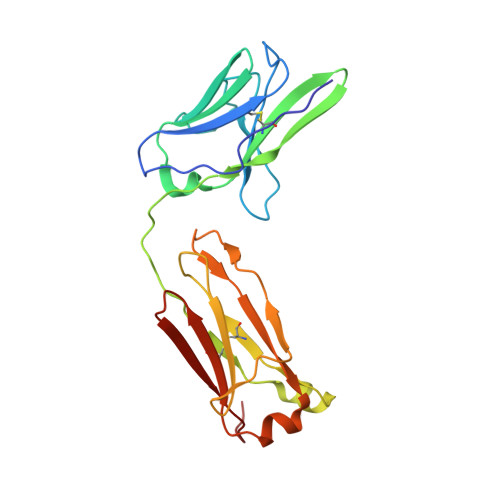
Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site.
Wen, X., Krause, J.C., Leser, G.P., Cox, R.G., Lamb, R.A., Williams, J.V., Crowe, J.E., Jardetzky, T.S.(2012) Nat Struct Mol Biol 19: 461-463
- PubMed: 22388735
- DOI: https://doi.org/10.1038/nsmb.2250
- Primary Citation of Related Structures:
4DAG - PubMed Abstract:
Human metapneumovirus and respiratory syncytial virus cause lower respiratory tract infections. The virus fusion (F) glycoprotein promotes membrane fusion by refolding from a metastable pre-fusion to a stable post-fusion conformation. F is also a major target of the neutralizing antibody response. Here we show that a potently neutralizing anti-human metapneumovirus antibody (DS7) binds a structurally invariant domain of F, revealing a new epitope that could be targeted in vaccine development.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, California, USA.