The Structure of the Cysteine Protease and Lectin-Like Domains of Cwp84, a Surface Layer-Associated Protein from Clostridium Difficile
Bradshaw, W.J., Kirby, J.M., Thiyagarajan, N., Chambers, C.J., Davies, A.H., Roberts, A.K., Shone, C.C., Acharya, K.R.(2014) Acta Crystallogr D Biol Crystallogr 70: 1983
- PubMed: 25004975
- DOI: https://doi.org/10.1107/S1399004714009997
- Primary Citation of Related Structures:
4CI7 - PubMed Abstract:
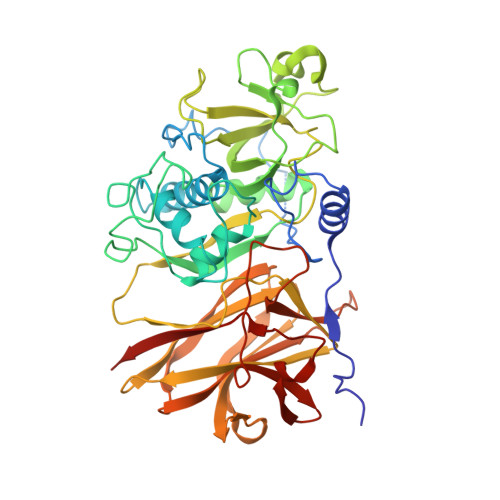
Clostridium difficile is a major problem as an aetiological agent for antibiotic-associated diarrhoea. The mechanism by which the bacterium colonizes the gut during infection is poorly understood, but undoubtedly involves a myriad of components present on the bacterial surface. The mechanism of C. difficile surface-layer (S-layer) biogenesis is also largely unknown but involves the post-translational cleavage of a single polypeptide (surface-layer protein A; SlpA) into low- and high-molecular-weight subunits by Cwp84, a surface-located cysteine protease. Here, the first crystal structure of the surface protein Cwp84 is described at 1.4 Å resolution and the key structural components are identified. The truncated Cwp84 active-site mutant (amino-acid residues 33-497; C116A) exhibits three regions: a cleavable propeptide and a cysteine protease domain which exhibits a cathepsin L-like fold followed by a newly identified putative carbohydrate-binding domain with a bound calcium ion, which is referred to here as a lectin-like domain. This study thus provides the first structural insights into Cwp84 and a strong base to elucidate its role in the C. difficile S-layer maturation mechanism.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Claverton Down, Bath BA2 7AY, England.