Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Weinert, C., Morger, D., Djekic, A., Mittl, P.R.E., Gruetter, M.G.(2015) Sci Rep 5: 10819
- PubMed: 26043233
- DOI: https://doi.org/10.1038/srep10819
- Primary Citation of Related Structures:
4CG4 - PubMed Abstract:
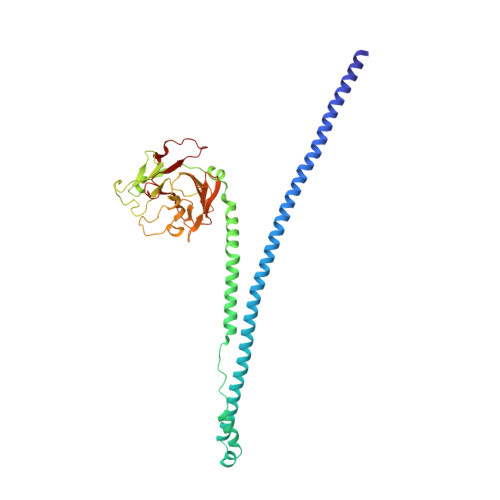
Many tripartite motif-containing (TRIM) proteins, comprising RING-finger, B-Box, and coiled-coil domains, carry additional B30.2 domains on the C-terminus of the TRIM motif and are considered to be pattern recognition receptors involved in the detection of higher order oligomers (e.g. viral capsid proteins). To investigate the spatial architecture of domains in TRIM proteins we determined the crystal structure of the TRIM20Δ413 fragment at 2.4 Å resolution. This structure comprises the central helical scaffold (CHS) and C-terminal B30.2 domains and reveals an anti-parallel arrangement of CHS domains placing the B-box domains 170 Å apart from each other. Small-angle X-ray scattering confirmed that the linker between CHS and B30.2 domains is flexible in solution. The crystal structure suggests an interaction between the B30.2 domain and an extended stretch in the CHS domain, which involves residues that are mutated in the inherited disease Familial Mediterranean Fever. Dimerization of B30.2 domains by means of the CHS domain is crucial for TRIM20 to bind pro-IL-1β in vitro. To exemplify how TRIM proteins could be involved in binding higher order oligomers we discuss three possible models for the TRIM5α/HIV-1 capsid interaction assuming different conformations of B30.2 domains.
Organizational Affiliation:
Department of Biochemistry, University Zürich, Winterthurerstrasse 190, 8057 Zürich, Switzerland.