How the Structure of the Large Subunit Controls Function in an Oxygen-Tolerant [Nife]-Hydrogenase.
Bowman, L., Flanagan, L., Fyfe, P.K., Parkin, A., Hunter, W.N., Sargent, F.(2014) Biochem J 458: 449
- PubMed: 24428762
- DOI: https://doi.org/10.1042/BJ20131520
- Primary Citation of Related Structures:
4C3O - PubMed Abstract:
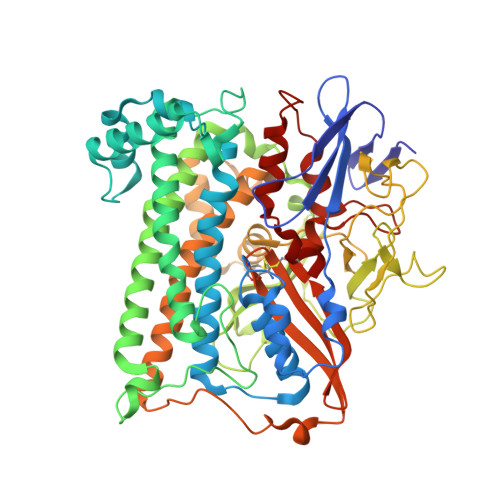
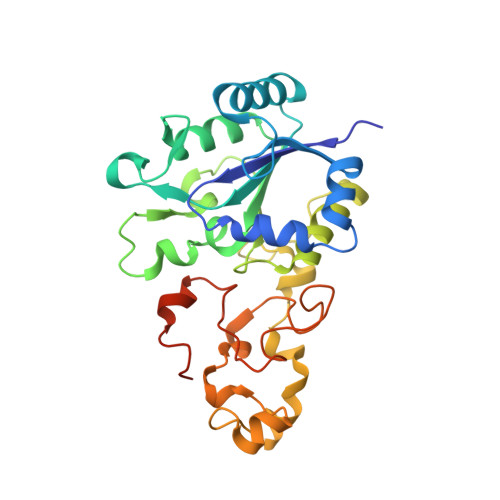
Salmonella enterica is an opportunistic pathogen that produces a [NiFe]-hydrogenase under aerobic conditions. In the present study, genetic engineering approaches were used to facilitate isolation of this enzyme, termed Hyd-5. The crystal structure was determined to a resolution of 3.2 Å and the hydro-genase was observed to comprise associated large and small subunits. The structure indicated that His229 from the large subunit was close to the proximal [4Fe-3S] cluster in the small subunit. In addition, His229 was observed to lie close to a buried glutamic acid (Glu73), which is conserved in oxygen-tolerant hydrogenases. His229 and Glu73 of the Hyd-5 large subunit were found to be important in both hydrogen oxidation activity and the oxygen-tolerance mechanism. Substitution of His229 or Glu73 with alanine led to a loss in the ability of Hyd-5 to oxidize hydrogen in air. Furthermore, the H229A variant was found to have lost the overpotential requirement for activity that is always observed with oxygen-tolerant [NiFe]-hydrogenases. It is possible that His229 has a role in stabilizing the super-oxidized form of the proximal cluster in the presence of oxygen, and it is proposed that Glu73could play a supporting role in fine-tuning the chemistry of His229 to enable this function.
Organizational Affiliation:
*Division of Molecular Microbiology, College of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland, U.K.