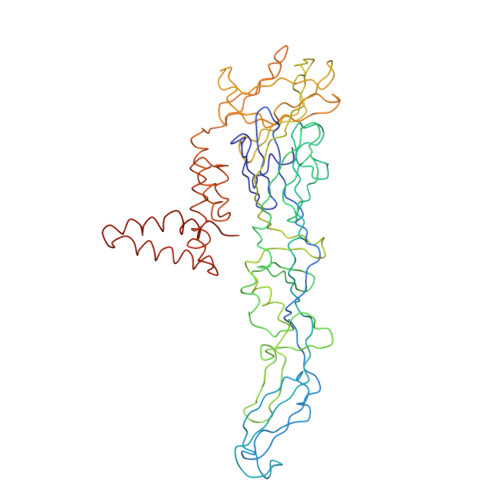
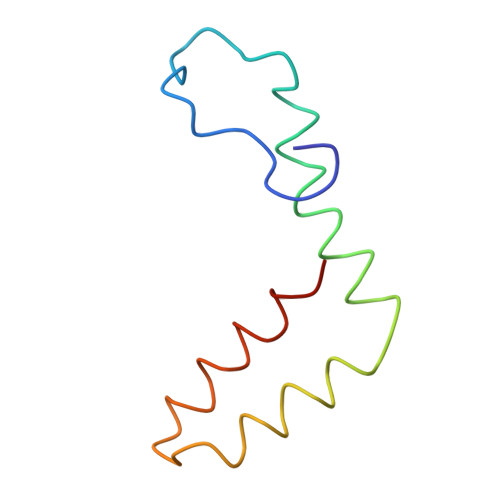
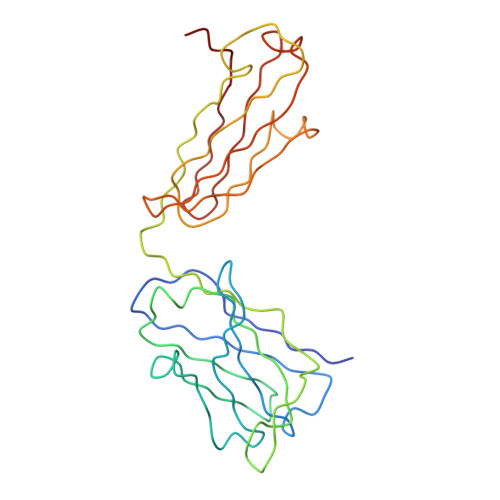
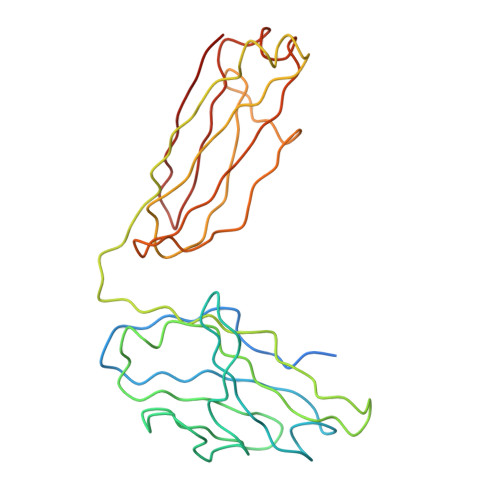
A Potent Anti-Dengue Human Antibody Preferentially Recognizes the Conformation of E Protein Monomers Assembled on the Virus Surface.
Fibriansah, G., Tan, J.L., Smith, S.A., De Alwis, A.R., Ng, T., Kostyuchenko, V.A., Ibarra, K.D., Wang, J., Harris, E., De Silva, A., Crowe, J.E.J., Lok, S.(2014) EMBO Mol Med 6: 358
- PubMed: 24421336
- DOI: https://doi.org/10.1002/emmm.201303404
- Primary Citation of Related Structures:
4C2I - PubMed Abstract:
Dengue virus (DENV), which consists of four serotypes (DENV1-4), infects over 400 million people annually. Previous studies have indicated most human monoclonal antibodies (HMAbs) from dengue patients are cross-reactive and poorly neutralizing. Rare neutralizing HMAbs are usually serotype-specific and bind to quaternary structure-dependent epitopes. We determined the structure of DENV1 complexed with Fab fragments of a highly potent HMAb 1F4 to 6 Å resolution by cryo-EM. Although HMAb 1F4 appeared to bind to virus and not E proteins in ELISAs in the previous study, our structure showed that the epitope is located within an envelope (E) protein monomer, and not across neighboring E proteins. The Fab molecules bind to domain I (DI), and DI-DII hinge of the E protein. We also showed that HMAb 1F4 can neutralize DENV at different stages of viral entry in a cell type and receptor dependent manner. The structure reveals the mechanism by which this potent and specific antibody blocks viral infection.
Organizational Affiliation:
Program in Emerging Infectious Diseases, Duke-NUS Graduate Medical School, Singapore City, Singapore.