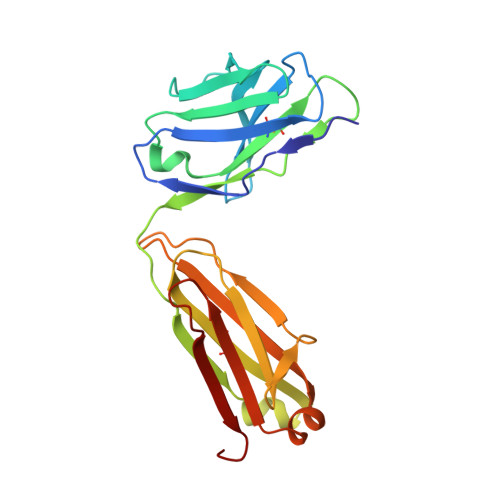
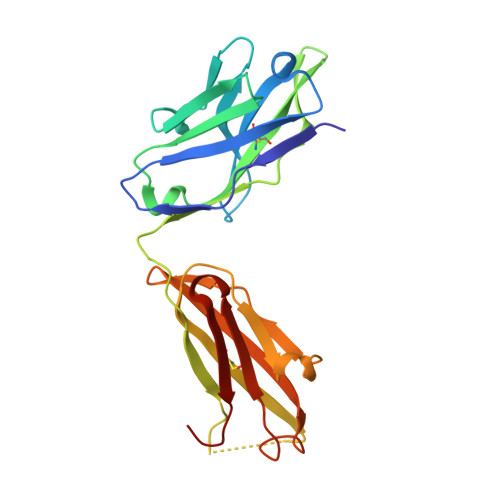
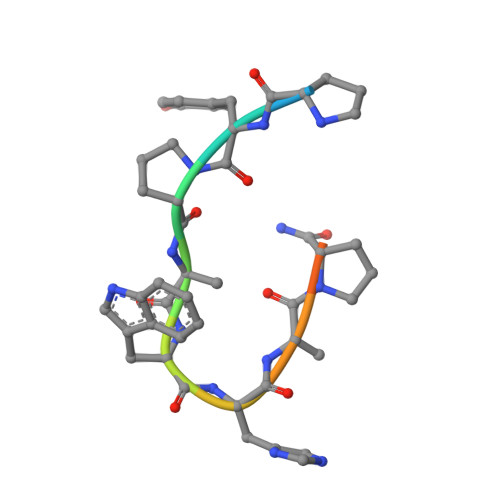
Adjustable Locks and Flexible Keys: Plasticity of Epitope-Paratope Interactions in Germline Antibodies.
Khan, T., Salunke, D.M.(2014) J Immunol 192: 5398
- PubMed: 24790145
- DOI: https://doi.org/10.4049/jimmunol.1302143
- Primary Citation of Related Structures:
4BH7, 4BH8 - PubMed Abstract:
Ag recognition by independent primary Abs against a small flexible Ag with overlapping epitopes was analyzed to address the determinants of Ag specificity during the initial encounter. Crystal structures of two distinct dodecapeptide Ags, GDPRPSYISHLL and PPYPAWHAPGNI, in complex with the germline mAb 36-65 were determined and compared with the structures of the same Ags bound to another independent germline mAb, BBE6.12H3. For each peptide Ag, the two germline mAbs recognized overlapping epitopes, but in different topologies. The peptide structures differed, and the two paratopes attained discrete conformations, leading to different surface topologies, in a mode that can be described as adjustable locks and flexible keys. This is in contrast to mature mAbs, in which conformational convergence of different paratopes while binding to a common epitope in a similar conformation has been reported. These results suggest that the primary immune receptor repertoire is highly versatile as compared with its mature counterpart. Germline and mature mAbs adopt distinct mechanisms for recognizing a flexible epitope. Whereas conservation of conformational repertoire is a key characteristic of mature mAbs achieved through affinity maturation, the germline mAbs, at the initial stages of Ag encounter, maintain substantial plasticity, accommodating a broad specificity repertoire.
Organizational Affiliation:
National Institute of Immunology, New Delhi 110067, India; and.