Capsid Structure and its Stability at the Late Stages of Bacteriophage Spp1 Assembly.
White, H.E., Sherman, M.B., Brasiles, S., Jacquet, E., Seavers, P., Tavares, P., Orlova, E.V.(2012) J Virol 86: 6768
- PubMed: 22514336
- DOI: https://doi.org/10.1128/JVI.00412-12
- Primary Citation of Related Structures:
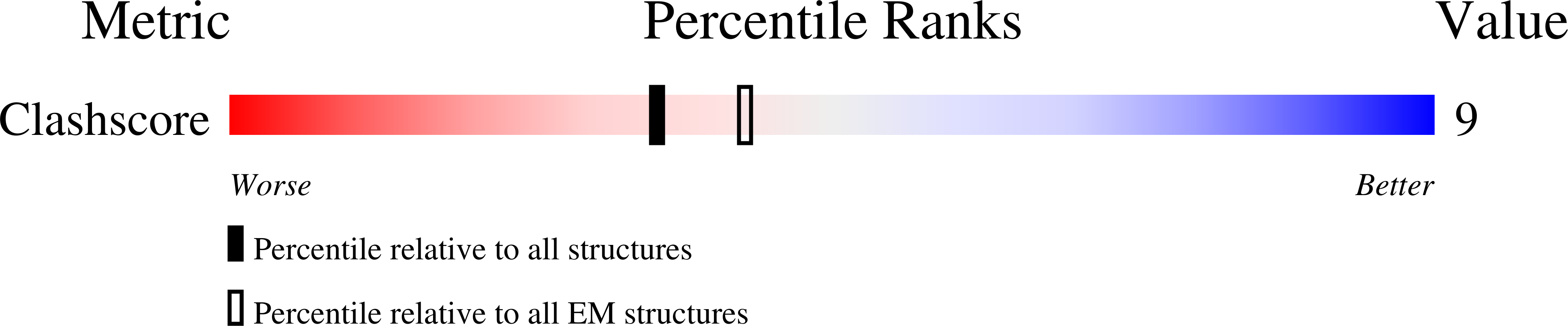
4AN5 - PubMed Abstract:
The structure of the bacteriophage SPP1 capsid was determined at subnanometer resolution by cryo-electron microscopy and single-particle analysis. The icosahedral capsid is composed of the major capsid protein gp13 and the auxiliary protein gp12, which are organized in a T=7 lattice. DNA is arranged in layers with a distance of ~24.5 Å. gp12 forms spikes that are anchored at the center of gp13 hexamers. In a gp12-deficient mutant, the centers of hexamers are closed by loops of gp13 coming together to protect the SPP1 genome from the outside environment. The HK97-like fold was used to build a pseudoatomic model of gp13. Its structural organization remains unchanged upon tail binding and following DNA release. gp13 exhibits enhanced thermostability in the DNA-filled capsid. A remarkable convergence between the thermostability of the capsid and those of the other virion components was found, revealing that the overall architecture of the SPP1 infectious particle coevolved toward high robustness.
Organizational Affiliation:
Crystallography, Institute for Structural and Molecular Biology, Department of Biological Sciences, Birkbeck College, University of London, London, United Kingdom.