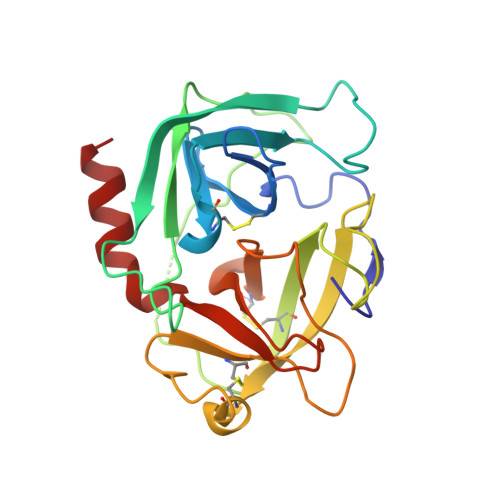
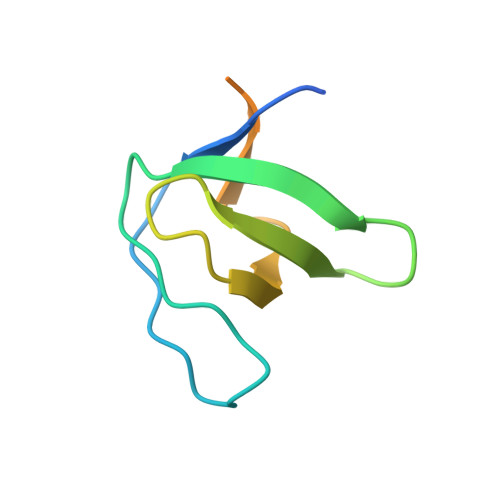
Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Schlatter, D., Brack, S., Banner, D.W., Batey, S., Benz, J., Bertschinger, J., Huber, W., Joseph, C., Rufer, A., Van Der Klooster, A., Weber, M., Grabulovski, D., Hennig, M.(2012) MAbs 4: 497
- PubMed: 22653218
- DOI: https://doi.org/10.4161/mabs.20452
- Primary Citation of Related Structures:
4AFQ, 4AFS, 4AFU, 4AFZ, 4AG1, 4AG2 - PubMed Abstract:
The serine protease chymase (EC = 3.4.21.39) is expressed in the secretory granules of mast cells, which are important in allergic reactions. Fynomers, which are binding proteins derived from the Fyn SH3 domain, were generated against human chymase to produce binding partners to facilitate crystallization, structure determination and structure-based drug discovery, and to provide inhibitors of chymase for therapeutic applications. The best Fynomer was found to bind chymase with a KD of 0.9 nM and koff of 6.6x10 (-4) s (-1) , and to selectively inhibit chymase activity with an IC 50 value of 2 nM. Three different Fynomers were co-crystallized with chymase in 6 different crystal forms overall, with diffraction quality in the range of 2.25 to 1.4 Å resolution, which is suitable for drug design efforts. The X-ray structures show that all Fynomers bind to the active site of chymase. The conserved residues Arg15-Trp16-Thr17 in the RT-loop of the chymase binding Fynomers provide a tight interaction, with Trp16 pointing deep into the S1 pocket of chymase. These results confirm the suitability of Fynomers as research tools to facilitate protein crystallization, as well as for the development of assays to investigate the biological mechanism of targets. Finally, their highly specific inhibitory activity and favorable molecular properties support the use of Fynomers as potential therapeutic agents.
Organizational Affiliation:
F. Hoffmann-La Roche Ltd, Pharma Research & Early Development, Discovery Technologies, Basel, Switzerland.