Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Luo, X., Wasilko, D.J., Liu, Y., Sun, J., Wu, X., Luo, Z.Q., Mao, Y.(2015) PLoS Pathog 11: e1004965-e1004965
- PubMed: 26067986
- DOI: https://doi.org/10.1371/journal.ppat.1004965
- Primary Citation of Related Structures:
4ZUZ - PubMed Abstract:
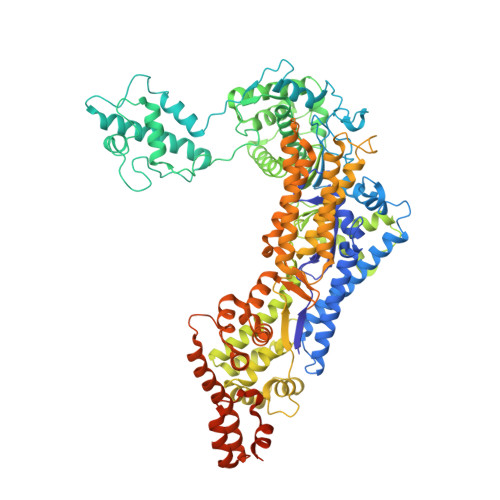
The opportunistic intracellular pathogen Legionella pneumophila is the causative agent of Legionnaires' disease. L. pneumophila delivers nearly 300 effector proteins into host cells for the establishment of a replication-permissive compartment known as the Legionella-containing vacuole (LCV). SidC and its paralog SdcA are two effectors that have been shown to anchor on the LCV via binding to phosphatidylinositol-4-phosphate [PI(4)P] to facilitate the recruitment of ER proteins to the LCV. We recently reported that the N-terminal SNL (SidC N-terminal E3 Ligase) domain of SidC is a ubiquitin E3 ligase, and its activity is required for the recruitment of ER proteins to the LCV. Here we report the crystal structure of SidC (1-871). The structure reveals that SidC contains four domains that are packed into an arch-like shape. The P4C domain (PI(4)P binding of SidC) comprises a four α-helix bundle and covers the ubiquitin ligase catalytic site of the SNL domain. Strikingly, a pocket with characteristic positive electrostatic potentials is formed at one end of this bundle. Liposome binding assays of the P4C domain further identified the determinants of phosphoinositide recognition and membrane interaction. Interestingly, we also found that binding with PI(4)P stimulates the E3 ligase activity, presumably due to a conformational switch induced by PI(4)P from a closed form to an open active form. Mutations of key residues involved in PI(4)P binding significantly reduced the association of SidC with the LCV and abolished its activity in the recruitment of ER proteins and ubiquitin signals, highlighting that PI(4)P-mediated targeting of SidC is critical to its function in the remodeling of the bacterial phagosome membrane. Finally, a GFP-fusion with the P4C domain was demonstrated to be specifically localized to PI(4)P-enriched compartments in mammalian cells. This domain shows the potential to be developed into a sensitive and accurate PI(4)P probe in living cells.
Organizational Affiliation:
Weill Institute for Cell and Molecular Biology and Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York, United States of America.