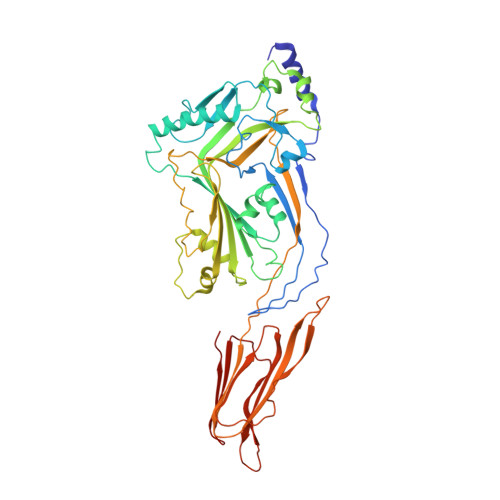
Crystal structure of Streptococcus pneumoniae pneumolysin provides key insights into early steps of pore formation.
Lawrence, S.L., Feil, S.C., Morton, C.J., Farrand, A.J., Mulhern, T.D., Gorman, M.A., Wade, K.R., Tweten, R.K., Parker, M.W.(2015) Sci Rep 5: 14352-14352
- PubMed: 26403197
- DOI: https://doi.org/10.1038/srep14352
- Primary Citation of Related Structures:
4ZGH - PubMed Abstract:
Pore-forming proteins are weapons often used by bacterial pathogens to breach the membrane barrier of target cells. Despite their critical role in infection important structural aspects of the mechanism of how these proteins assemble into pores remain unknown. Streptococcus pneumoniae is the world's leading cause of pneumonia, meningitis, bacteremia and otitis media. Pneumolysin (PLY) is a major virulence factor of S. pneumoniae and a target for both small molecule drug development and vaccines. PLY is a member of the cholesterol-dependent cytolysins (CDCs), a family of pore-forming toxins that form gigantic pores in cell membranes. Here we present the structure of PLY determined by X-ray crystallography and, in solution, by small-angle X-ray scattering. The crystal structure reveals PLY assembles as a linear oligomer that provides key structural insights into the poorly understood early monomer-monomer interactions of CDCs at the membrane surface.
Organizational Affiliation:
ACRF Rational Drug Discovery Centre, St. Vincent's Institute of Medical Research, Fitzroy, Victoria 3065, Australia.