A novel inert crystal delivery medium for serial femtosecond crystallography.
Conrad, C.E., Basu, S., James, D., Wang, D., Schaffer, A., Roy-Chowdhury, S., Zatsepin, N.A., Aquila, A., Coe, J., Gati, C., Hunter, M.S., Koglin, J.E., Kupitz, C., Nelson, G., Subramanian, G., White, T.A., Zhao, Y., Zook, J., Boutet, S., Cherezov, V., Spence, J.C., Fromme, R., Weierstall, U., Fromme, P.(2015) IUCrJ 2: 421-430
- PubMed: 26177184
- DOI: https://doi.org/10.1107/S2052252515009811
- Primary Citation of Related Structures:
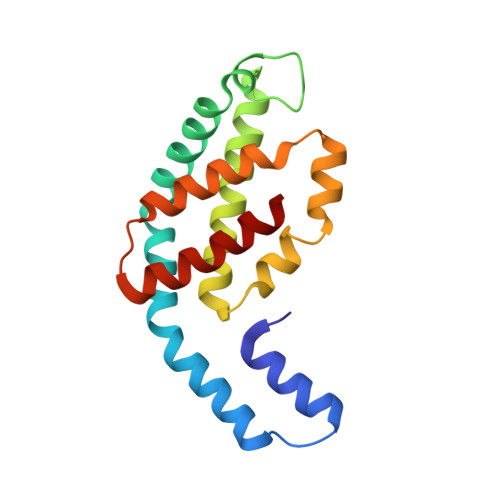
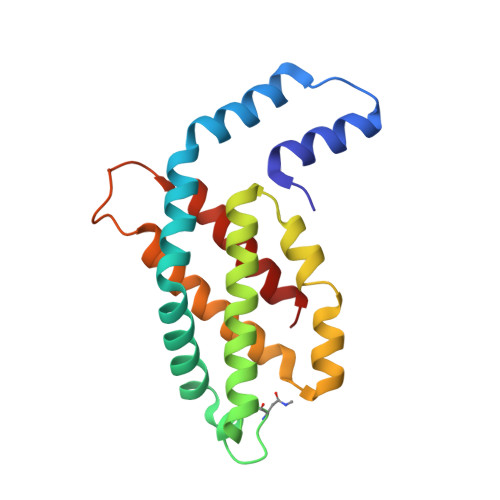
4Z8K - PubMed Abstract:
Serial femtosecond crystallography (SFX) has opened a new era in crystallo-graphy by permitting nearly damage-free, room-temperature structure determination of challenging proteins such as membrane proteins. In SFX, femtosecond X-ray free-electron laser pulses produce diffraction snapshots from nanocrystals and microcrystals delivered in a liquid jet, which leads to high protein consumption. A slow-moving stream of agarose has been developed as a new crystal delivery medium for SFX. It has low background scattering, is compatible with both soluble and membrane proteins, and can deliver the protein crystals at a wide range of temperatures down to 4°C. Using this crystal-laden agarose stream, the structure of a multi-subunit complex, phycocyanin, was solved to 2.5 Å resolution using 300 µg of microcrystals embedded into the agarose medium post-crystallization. The agarose delivery method reduces protein consumption by at least 100-fold and has the potential to be used for a diverse population of proteins, including membrane protein complexes.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Arizona State University , PO Box 871604, Tempe, AZ 85287-1604, USA ; Center for Applied Structural Discovery, The Biodesign Institute , PO Box 875001, Tempe, AZ 85287-5001, USA.