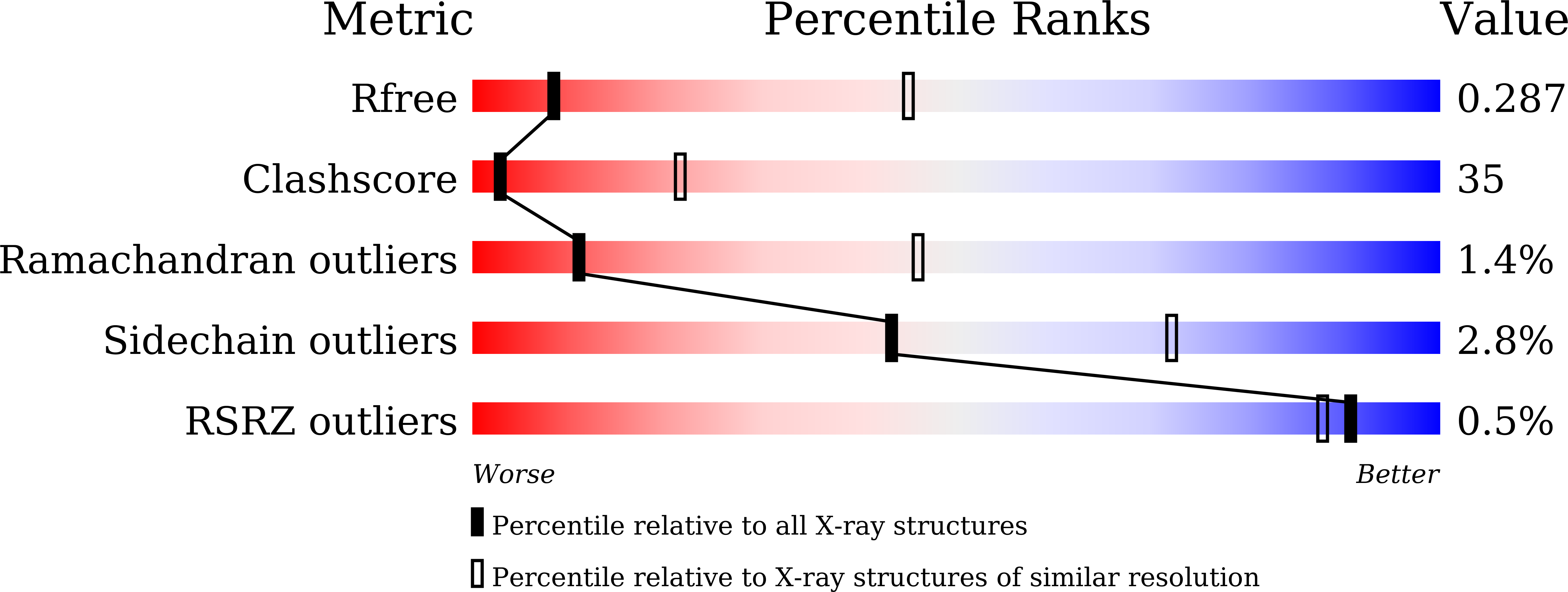
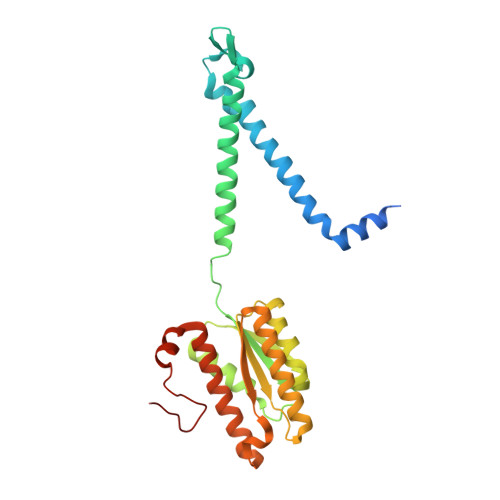
Mechanical coupling of the multiple structural elements of the large-conductance mechanosensitive channel during expansion
Li, J., Guo, J., Ou, X., Zhang, M., Li, Y., Liu, Z.(2015) Proc Natl Acad Sci U S A 112: 10726-10731
- PubMed: 26261325
- DOI: https://doi.org/10.1073/pnas.1503202112
- Primary Citation of Related Structures:
4Y7J, 4Y7K - PubMed Abstract:
The prokaryotic mechanosensitive channel of large conductance (MscL) is a pressure-relief valve protecting the cell from lysing during acute osmotic downshock. When the membrane is stretched, MscL responds to the increase of membrane tension and opens a nonselective pore to about 30 Å wide, exhibiting a large unitary conductance of ∼ 3 nS. A fundamental step toward understanding the gating mechanism of MscL is to decipher the molecular details of the conformational changes accompanying channel opening. By applying fusion-protein strategy and controlling detergent composition, we have solved the structures of an archaeal MscL homolog from Methanosarcina acetivorans trapped in the closed and expanded intermediate states. The comparative analysis of these two new structures reveals significant conformational rearrangements in the different domains of MscL. The large changes observed in the tilt angles of the two transmembrane helices (TM1 and TM2) fit well with the helix-pivoting model derived from the earlier geometric analyses based on the previous structures. Meanwhile, the periplasmic loop region transforms from a folded structure, containing an ω-shaped loop and a short β-hairpin, to an extended and partly disordered conformation during channel expansion. Moreover, a significant rotating and sliding of the N-terminal helix (N-helix) is coupled to the tilting movements of TM1 and TM2. The dynamic relationships between the N-helix and TM1/TM2 suggest that the N-helix serves as a membrane-anchored stopper that limits the tilts of TM1 and TM2 in the gating process. These results provide direct mechanistic insights into the highly coordinated movement of the different domains of the MscL channel when it expands.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China;