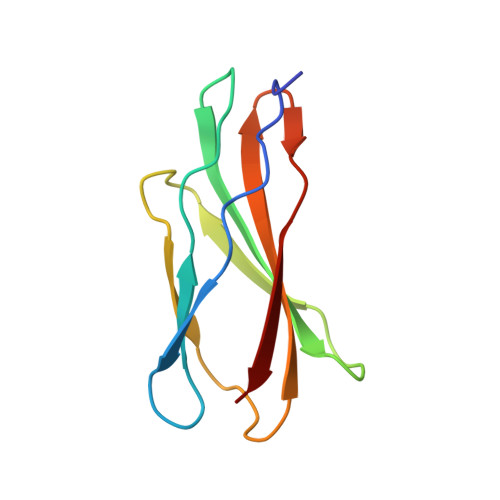
Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin alpha6beta4
Alonso-Garcia, N., Garcia-Rubio, I., Manso, J.A., Buey, R.M., Urien, H., Sonnenberg, A., Jeschke, G., de Pereda, J.M.(2015) Acta Crystallogr D Biol Crystallogr 71: 969-985
- PubMed: 25849406
- DOI: https://doi.org/10.1107/S1399004715002485
- Primary Citation of Related Structures:
4WTW, 4WTX - PubMed Abstract:
Integrin α6β4 is a major component of hemidesmosomes that mediate the stable anchorage of epithelial cells to the underlying basement membrane. Integrin α6β4 has also been implicated in cell proliferation and migration and in carcinoma progression. The third and fourth fibronectin type III domains (FnIII-3,4) of integrin β4 mediate binding to the hemidesmosomal proteins BPAG1e and BPAG2, and participate in signalling. Here, it is demonstrated that X-ray crystallography, small-angle X-ray scattering and double electron-electron resonance (DEER) complement each other to solve the structure of the FnIII-3,4 region. The crystal structures of the individual FnIII-3 and FnIII-4 domains were solved and the relative arrangement of the FnIII domains was elucidated by combining DEER with site-directed spin labelling. Multiple structures of the interdomain linker were modelled by Monte Carlo methods complying with DEER constraints, and the final structures were selected against experimental scattering data. FnIII-3,4 has a compact and cambered flat structure with an evolutionary conserved surface that is likely to correspond to a protein-interaction site. Finally, this hybrid method is of general application for the study of other macromolecules and complexes.
Organizational Affiliation:
Instituto de Biología Molecular y Celular del Cancer, Consejo Superior de Investigaciones Científicas - University of Salamanca, Campus Unamuno, 37007 Salamanca, Spain.