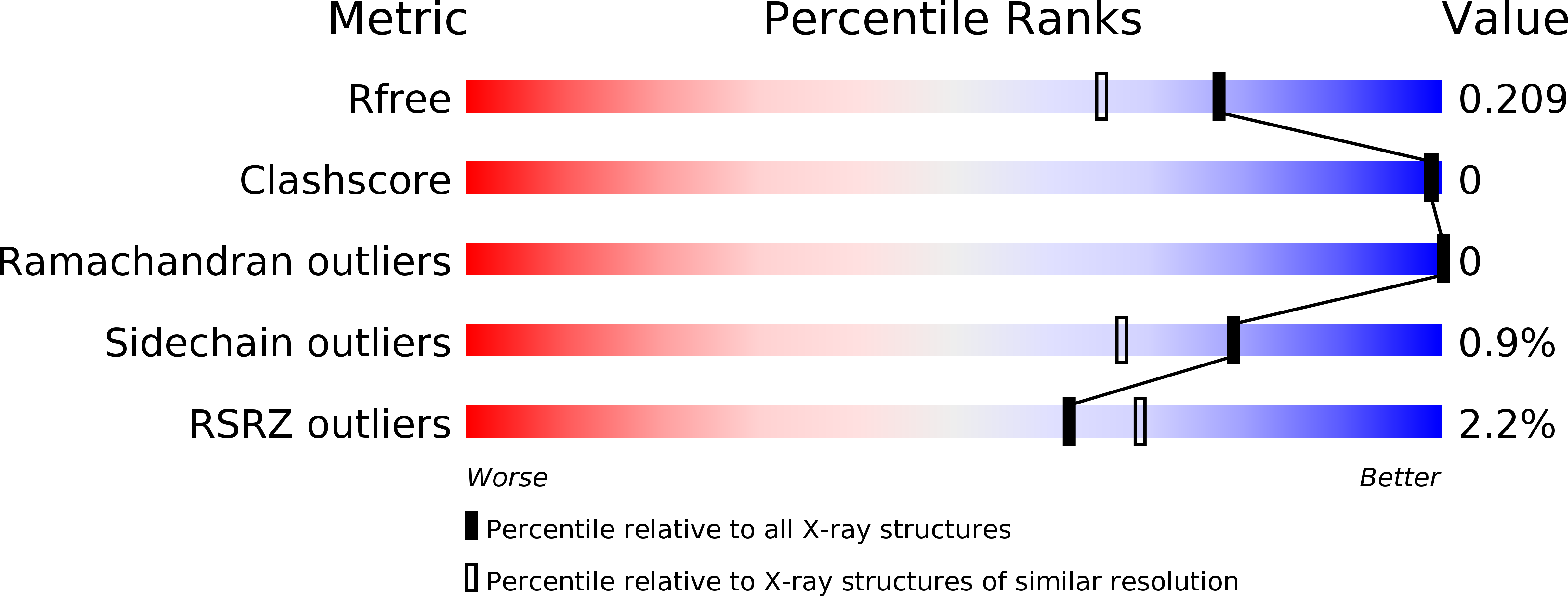
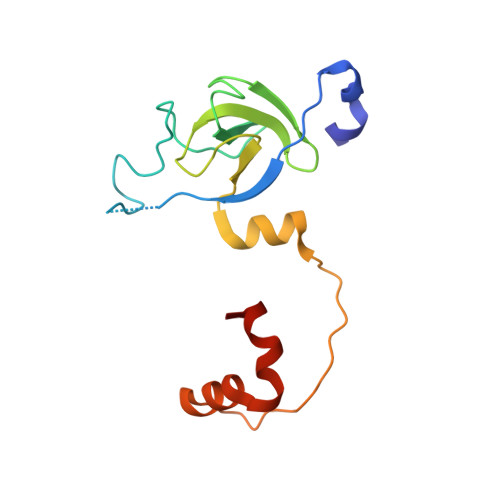
Structure of the stationary phase survival protein YuiC from B.subtilis.
Quay, D.H., Cole, A.R., Cryar, A., Thalassinos, K., Williams, M.A., Bhakta, S., Keep, N.H.(2015) BMC Struct Biol 15: 12-12
- PubMed: 26163297
- DOI: https://doi.org/10.1186/s12900-015-0039-z
- Primary Citation of Related Structures:
4WJT, 4WLI, 4WLK - PubMed Abstract:
Stationary phase survival proteins (Sps) were found in Firmicutes as having analogous domain compositions, and in some cases genome context, as the resuscitation promoting factors of Actinobacteria, but with a different putative peptidoglycan cleaving domain. The first structure of a Firmicute Sps protein YuiC from B. subtilis, is found to be a stripped down version of the cell-wall peptidoglycan hydrolase MltA. The YuiC structures are of a domain swapped dimer, although some monomer is also found in solution. The protein crystallised in the presence of pentasaccharide shows a 1,6-anhydrodisaccharide sugar product, indicating that YuiC cleaves the sugar backbone to form an anhydro product at least on lengthy incubation during crystallisation. The structural simplification of MltA in Sps proteins is analogous to that of the resuscitation promoting factor domains of Actinobacteria, which are stripped down versions of lysozyme and soluble lytic transglycosylase proteins.
Organizational Affiliation:
Institute for Structural and Molecular Biology, Crystallography, Department of Biological Sciences, Birkbeck University of London, Malet Street, London, WC1E 7HX, UK. d.quay@mail.cryst.bbk.ac.uk.