Essential role of the C-terminal helical domain in active site formation of selenoprotein MsrA from Clostridium oremlandii
Lee, E.H., Lee, K., Hwang, K.Y., Kim, H.-Y.(2015) PLoS One 10: e0117836-e0117836
- PubMed: 25692691
- DOI: https://doi.org/10.1371/journal.pone.0117836
- Primary Citation of Related Structures:
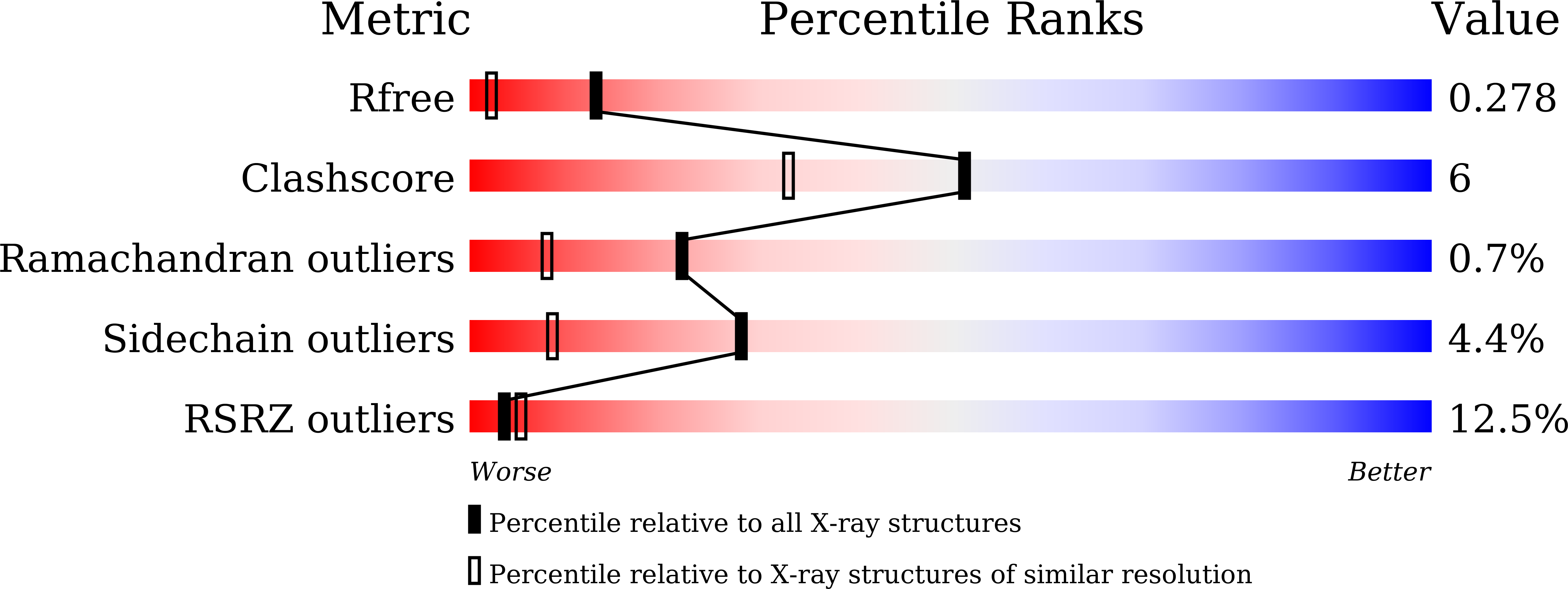
4W8C - PubMed Abstract:
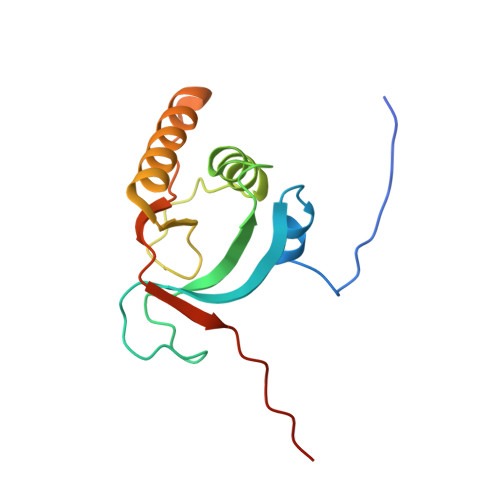
We previously determined the crystal structures of 1-Cys type selenoprotein MsrA from Clostridium oremlandii (CoMsrA). The overall structure of CoMsrA is unusual, consisting of two domains, the N-terminal catalytic domain and the C-terminal distinct helical domain which is absent from other known MsrA structures. Deletion of the helical domain almost completely abolishes the catalytic activity of CoMsrA. In this study, we determined the crystal structure of the helical domain-deleted (ΔH-domain) form of CoMsrA at a resolution of 1.76 Å. The monomer structure is composed of the central rolled mixed β-sheet surrounded by α-helices. However, there are significant conformational changes in the N- and C-termini and loop regions of the ΔH-domain protein relative to the catalytic domain structure of full-length CoMsrA. The active site structure in the ΔH-domain protein completely collapses, thereby causing loss of catalytic activity of the protein. Interestingly, dimer structures are observed in the crystal formed by N-terminus swapping between two molecules. The ΔH-domain protein primarily exists as a dimer in solution, whereas the full-length CoMsrA exists as a monomer. Collectively, this study provides insight into the structural basis of the essential role of the helical domain of CoMsrA in its catalysis.
Organizational Affiliation:
Division of Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul, Republic of Korea.