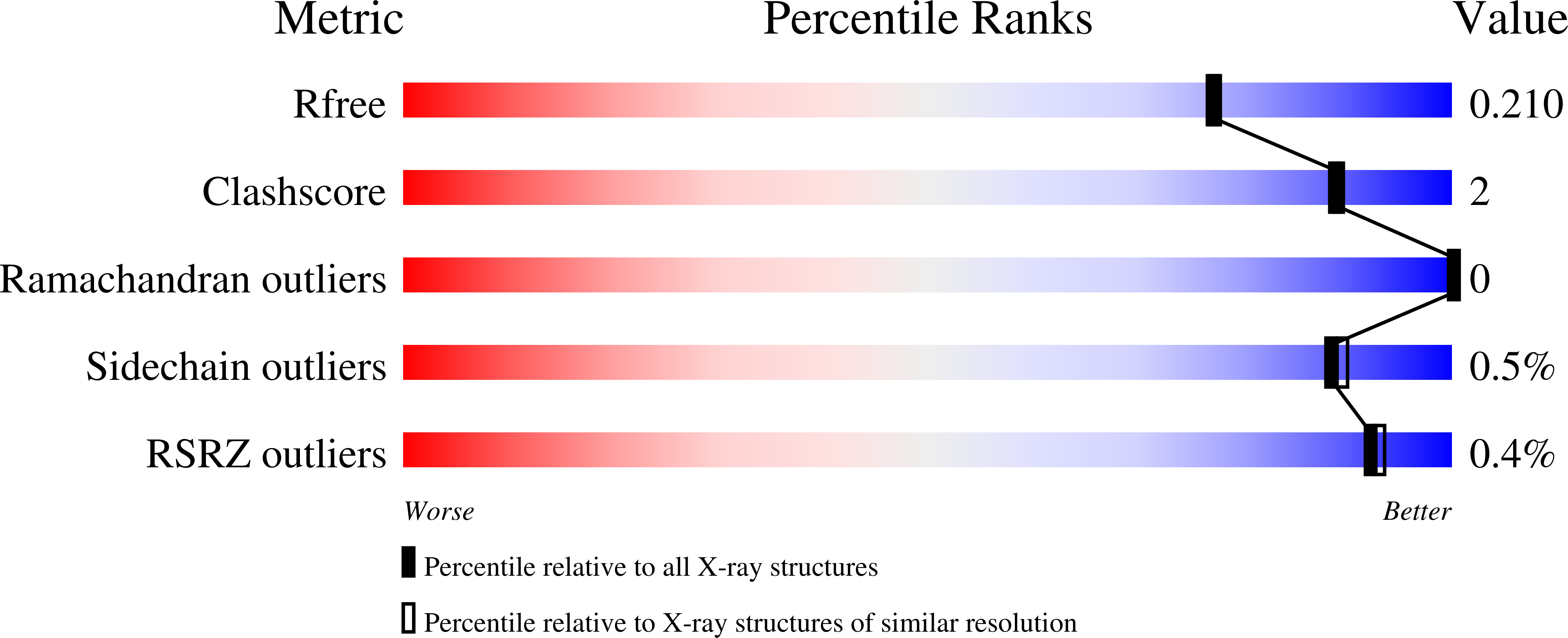
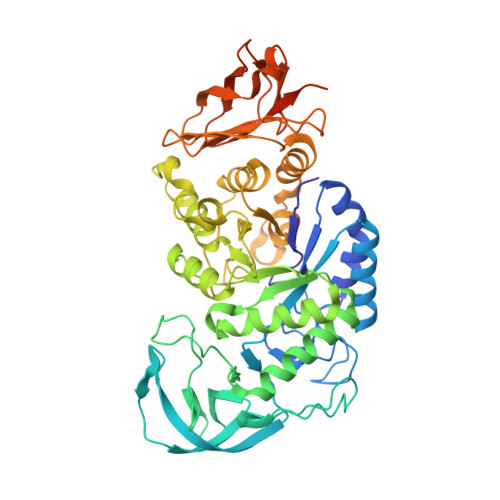
Three-Dimensional Structure of a Variant `Termamyl-Like' Geobacillus Stearothermophilus Alpha-Amylase at 1.9 A Resolution
Offen, W.A., Viksoe-Nielsen, A., Borchert, T.V., Wilson, K.S., Davies, G.J.(2015) Acta Crystallogr Sect F Struct Biol Cryst Commun 71: 66
- PubMed: 25615972
- DOI: https://doi.org/10.1107/S2053230X14026508
- Primary Citation of Related Structures:
4UZU - PubMed Abstract:
The enzyme-catalysed degradation of starch is central to many industrial processes, including sugar manufacture and first-generation biofuels. Classical biotechnological platforms involve steam explosion of starch followed by the action of endo-acting glycoside hydrolases termed α-amylases and then exo-acting α-glucosidases (glucoamylases) to yield glucose, which is subsequently processed. A key enzymatic player in this pipeline is the `Termamyl' class of bacterial α-amylases and designed/evolved variants thereof. Here, the three-dimensional structure of one such Termamyl α-amylase variant based upon the parent Geobacillus stearothermophilus α-amylase is presented. The structure has been solved at 1.9 Å resolution, revealing the classical three-domain fold stabilized by Ca2+ and a Ca2+-Na+-Ca2+ triad. As expected, the structure is similar to the G. stearothermophilus α-amylase but with main-chain deviations of up to 3 Å in some regions, reflecting both the mutations and differing crystal-packing environments.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, The University of York, York YO10 5DD, England.