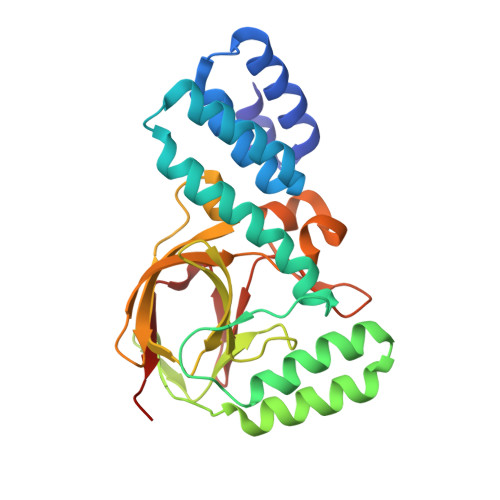
Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
Forster, A., Planamente, S., Manoli, E., Lossi, N.S., Freemont, P.S., Filloux, A.(2014) RNA 20: 1955
- PubMed: 25305017
- DOI: https://doi.org/10.1074/jbc.M114.600510
- Primary Citation of Related Structures:
4UQW, 4UQX, 4UQY, 4UQZ - PubMed Abstract:
The type VI secretion system (T6SS) is a bacterial nanomachine for the transport of effector molecules into prokaryotic and eukaryotic cells. It involves the assembly of a tubular structure composed of TssB and TssC that is similar to the tail sheath of bacteriophages. The sheath contracts to provide the energy needed for effector delivery. The AAA(+) ATPase ClpV disassembles the contracted sheath, which resets the systems for reassembly of an extended sheath that is ready to fire again. This mechanism is crucial for T6SS function. In Vibrio cholerae, ClpV binds the N terminus of TssC within a hydrophobic groove. In this study, we resolved the crystal structure of the N-terminal domain of Pseudomonas aeruginosa ClpV1 and observed structural alterations in the hydrophobic groove. The modification in the ClpV1 groove is matched by a change in the N terminus of TssC, suggesting the existence of distinct T6SS classes. An accessory T6SS component, TagJ/HsiE, exists predominantly in one of the classes. Using bacterial two-hybrid approaches, we showed that the P. aeruginosa homolog HsiE1 interacts strongly with ClpV1. We then resolved the crystal structure of HsiE1 in complex with the N terminus of HsiB1, a TssB homolog and component of the contractile sheath. Phylogenetic analysis confirmed that these differences distinguish T6SS classes that resulted from a functional co-evolution between TssB, TssC, TagJ/HsiE, and ClpV. The interaction of TagJ/HsiE with the sheath as well as with ClpV suggests an alternative mode of disassembly in which HsiE recruits the ATPase to the sheath.
Organizational Affiliation:
From the Centre for Structural Biology and.