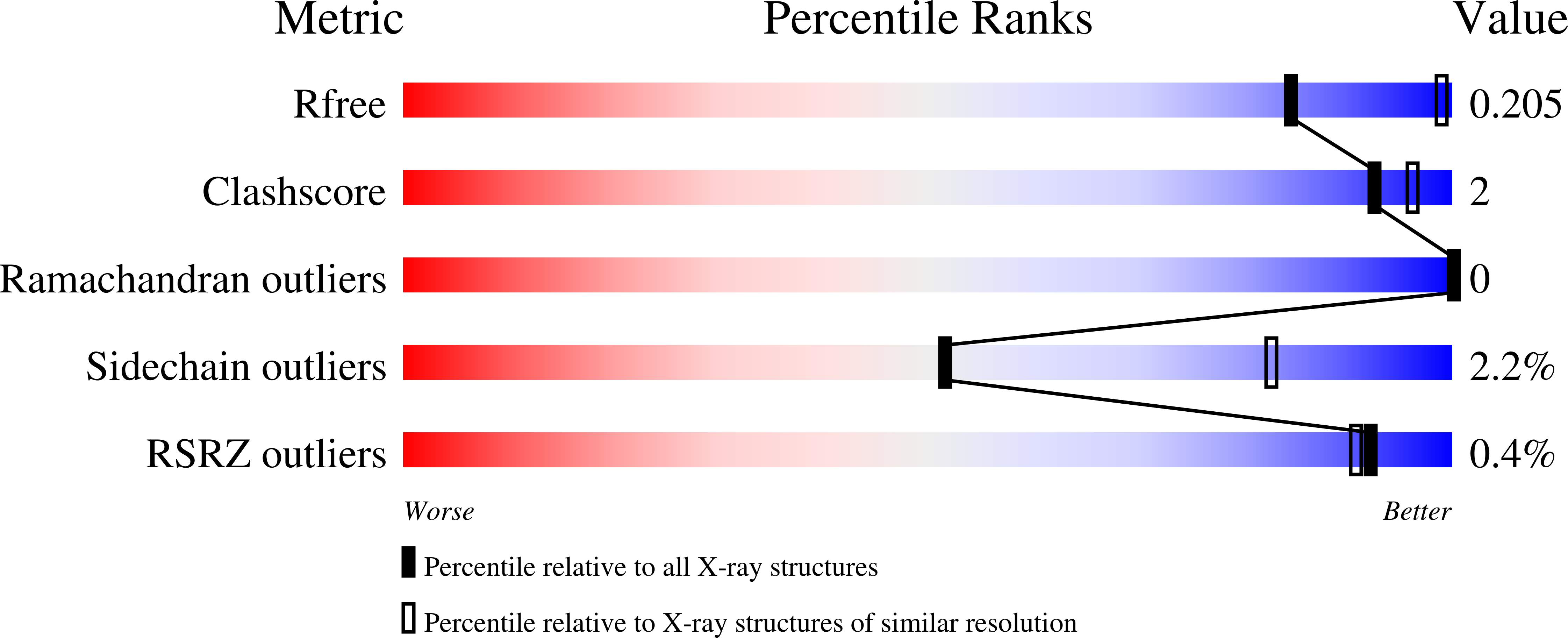
Action at a Distance: MUTATIONS OF PERIPHERAL RESIDUES TRANSFORM RAPID REVERSIBLE INHIBITORS TO SLOW, TIGHT BINDERS OF CYCLOOXYGENASE-2.
Blobaum, A.L., Xu, S., Rowlinson, S.W., Duggan, K.C., Banerjee, S., Kudalkar, S.N., Birmingham, W.R., Ghebreselasie, K., Marnett, L.J.(2015) J Biol Chem 290: 12793-12803
- PubMed: 25825493
- DOI: https://doi.org/10.1074/jbc.M114.635987
- Primary Citation of Related Structures:
4RRW, 4RRX, 4RRY, 4RRZ, 4RS0 - PubMed Abstract:
Cyclooxygenase enzymes (COX-1 and COX-2) catalyze the conversion of arachidonic acid to prostaglandin G2. The inhibitory activity of rapid, reversible COX inhibitors (ibuprofen, naproxen, mefenamic acid, and lumiracoxib) demonstrated a significant increase in potency and time dependence of inhibition against double tryptophan murine COX-2 mutants at the 89/90 and 89/119 positions. In contrast, the slow, time-dependent COX inhibitors (diclofenac, indomethacin, and flurbiprofen) were unaffected by those mutations. Further mutagenesis studies suggested that mutation at position 89 was principally responsible for the changes in inhibitory potency of rapid, reversible inhibitors, whereas mutation at position 90 may exert some effect on the potency of COX-2-selective diarylheterocycle inhibitors; no effect was observed with mutation at position 119. Several crystal structures with or without NSAIDs indicated that placement of a bulky residue at position 89 caused a closure of a gap at the lobby, and alteration of histidine to tryptophan at position 90 changed the electrostatic profile of the side pocket of COX-2. Thus, these two residues, especially Val-89 at the lobby region, are crucial for the entrance and exit of some NSAIDs from the COX active site.
Organizational Affiliation:
From the A. B. Hancock, Jr. Memorial Laboratory for Cancer Research, Departments of Biochemistry, Chemistry, and Pharmacology, Vanderbilt Institute of Chemical Biology, Center in Molecular Toxicology, Vanderbilt-Ingram Cancer Center, Vanderbilt University School of Medicine, Nashville Tennessee 37232.