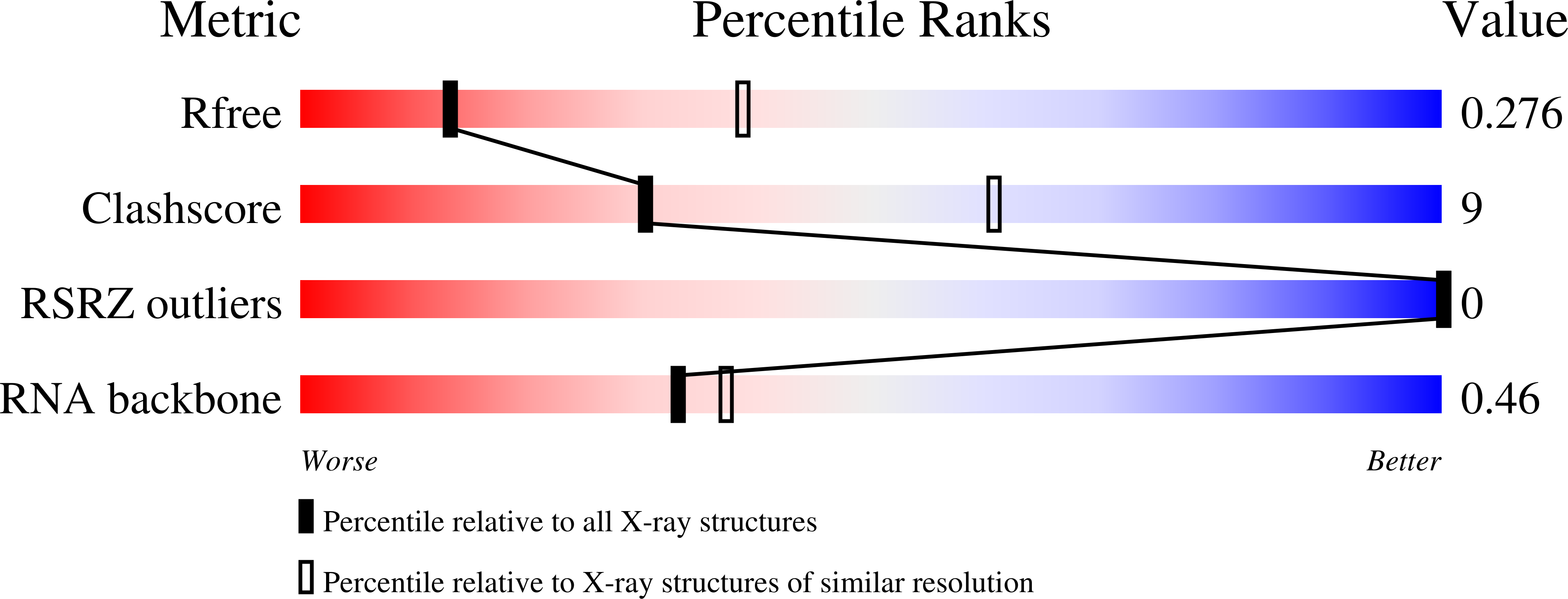
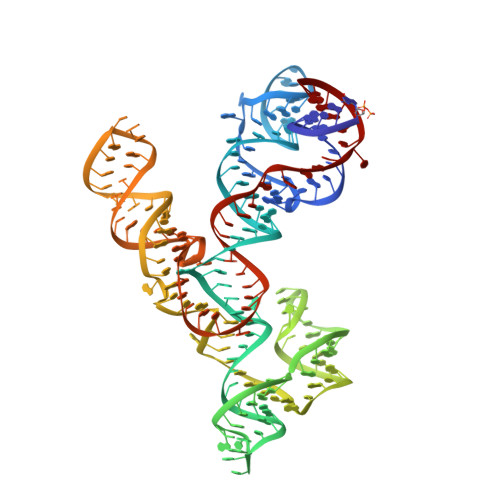
Crystal structure of the Varkud satellite ribozyme.
Suslov, N.B., DasGupta, S., Huang, H., Fuller, J.R., Lilley, D.M., Rice, P.A., Piccirilli, J.A.(2015) Nat Chem Biol 11: 840-846
- PubMed: 26414446
- DOI: https://doi.org/10.1038/nchembio.1929
- Primary Citation of Related Structures:
4R4P, 4R4V - PubMed Abstract:
The Varkud satellite (VS) ribozyme mediates rolling-circle replication of a plasmid found in the Neurospora mitochondrion. We report crystal structures of this ribozyme from Neurospora intermedia at 3.1 Å resolution, which revealed an intertwined dimer formed by an exchange of substrate helices. In each protomer, an arrangement of three-way helical junctions organizes seven helices into a global fold that creates a docking site for the substrate helix of the other protomer, resulting in the formation of two active sites in trans. This mode of RNA-RNA association resembles the process of domain swapping in proteins and has implications for RNA regulation and evolution. Within each active site, adenine and guanine nucleobases abut the scissile phosphate, poised to serve direct roles in catalysis. Similarities to the active sites of the hairpin and hammerhead ribozymes highlight the functional importance of active-site features, underscore the ability of RNA to access functional architectures from distant regions of sequence space, and suggest convergent evolution.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois, USA.