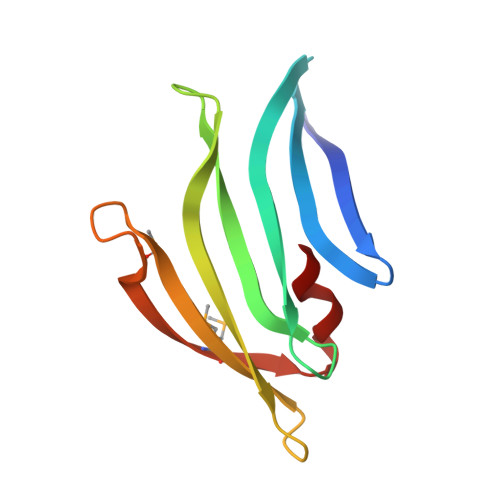
Structures of single-layer beta-sheet proteins evolved from beta-hairpin repeats.
Xu, Q., Biancalana, M., Grant, J.C., Chiu, H.J., Jaroszewski, L., Knuth, M.W., Lesley, S.A., Godzik, A., Elsliger, M.A., Deacon, A.M., Wilson, I.A.(2019) Protein Sci 28: 1676-1689
- PubMed: 31306512
- DOI: https://doi.org/10.1002/pro.3683
- Primary Citation of Related Structures:
3MSW, 4R03, 4R8O - PubMed Abstract:
Free-standing single-layer β-sheets are extremely rare in naturally occurring proteins, even though β-sheet motifs are ubiquitous. Here we report the crystal structures of three homologous, single-layer, anti-parallel β-sheet proteins, comprised of three or four twisted β-hairpin repeats. The structures reveal that, in addition to the hydrogen bond network characteristic of β-sheets, additional hydrophobic interactions mediated by small clusters of residues adjacent to the turns likely play a significant role in the structural stability and compensate for the lack of a compact hydrophobic core. These structures enabled identification of a family of secreted proteins that are broadly distributed in bacteria from the human gut microbiome and are putatively involved in the metabolism of complex carbohydrates. A conserved surface patch, rich in solvent-exposed tyrosine residues, was identified on the concave surface of the β-sheet. These new modular single-layer β-sheet proteins may serve as a new model system for studying folding and design of β-rich proteins.
Organizational Affiliation:
Joint Center for Structural Genomics, www.jcsg.org.