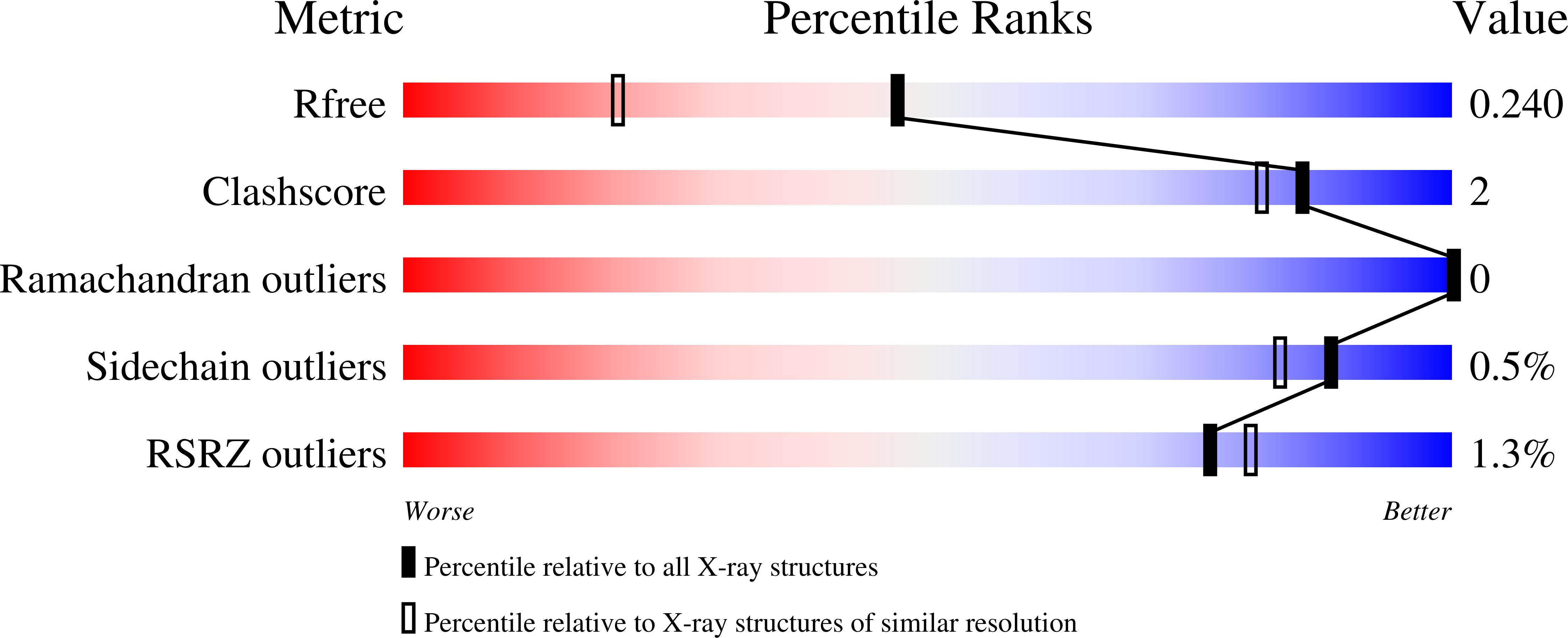
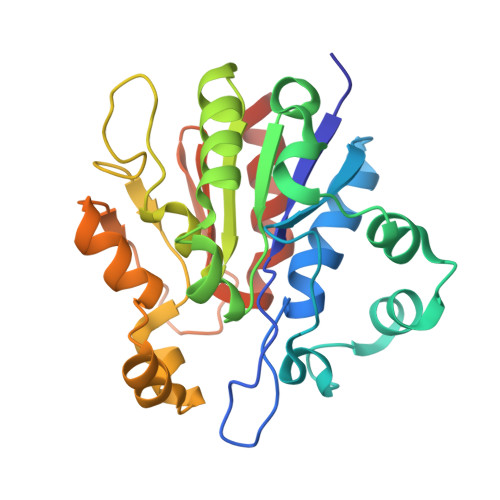
The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Wilson, M.A., Amour, S.T., Collins, J.L., Ringe, D., Petsko, G.A.(2004) Proc Natl Acad Sci U S A 101: 1531-1536
- PubMed: 14745011
- DOI: https://doi.org/10.1073/pnas.0308089100
- Primary Citation of Related Structures:
4QYX - PubMed Abstract:
The yeast gene YDR533C encodes a protein belonging to the DJ-1/ThiJ/PfpI superfamily. This family includes the human protein DJ-1, which is mutated in autosomal recessive early-onset Parkinson's disease. The function of DJ-1 and its yeast homologue YDR533Cp is unknown. We report here the crystal structure of YDR533Cp at 1.8-A resolution. The structure indicates that the closest relative to YDR533Cp is the Escherichia coli heat shock protein Hsp31 (YedU), which has both chaperone and protease activity. As expected, the overall fold of the core domain of YDR533Cp is also similar to that of DJ-1 and the bacterial protease PfpI. YDR533Cp contains a possible catalytic triad analogous to that of Hsp31 and an additional domain that is present in Hsp31 but is not seen in DJ-1 and other members of the family. The cysteine in this triad (Cys-138) is oxidized in this crystal structure, similar to modifications seen in the corresponding cysteine in the crystal structure of DJ-1. YDR533Cp appears to be a dimer both in solution and the crystal, but this dimer is formed by a different interface than that found in Hsp31 or other members of the superfamily.
Organizational Affiliation:
Departments of Biochemistry and Chemistry, and Rosenstiel Basic Medical Sciences Research Center, Brandeis University, 415 South Street, MS 029, Waltham, MA 02454-9110, USA.