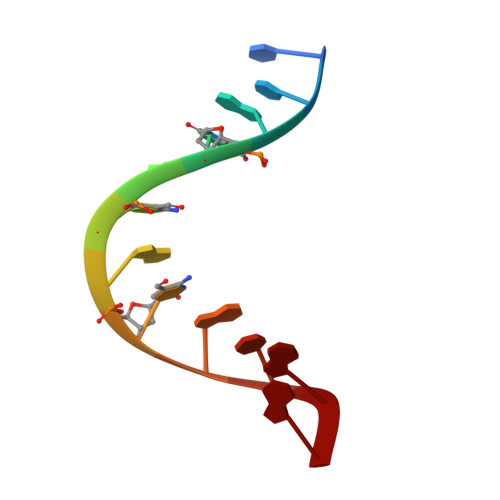
5-Formylcytosine alters the structure of the DNA double helix.
Raiber, E.A., Murat, P., Chirgadze, D.Y., Beraldi, D., Luisi, B.F., Balasubramanian, S.(2015) Nat Struct Mol Biol 22: 44-49
- PubMed: 25504322
- DOI: https://doi.org/10.1038/nsmb.2936
- Primary Citation of Related Structures:
4QKK - PubMed Abstract:
The modified base 5-formylcytosine (5fC) was recently identified in mammalian DNA and might be considered to be the 'seventh' base of the genome. This nucleotide has been implicated in active demethylation mediated by the base excision repair enzyme thymine DNA glycosylase. Genomics and proteomics studies have suggested an additional role for 5fC in transcription regulation through chromatin remodeling. Here we propose that 5fC might affect these processes through its effect on DNA conformation. Biophysical and structural analysis revealed that 5fC alters the structure of the DNA double helix and leads to a conformation unique among known DNA structures including those comprising other cytosine modifications. The 1.4-Å-resolution X-ray crystal structure of a DNA dodecamer comprising three 5fCpG sites shows how 5fC changes the geometry of the grooves and base pairs associated with the modified base, leading to helical underwinding.
Organizational Affiliation:
Department of Chemistry, University of Cambridge, Cambridge, UK.