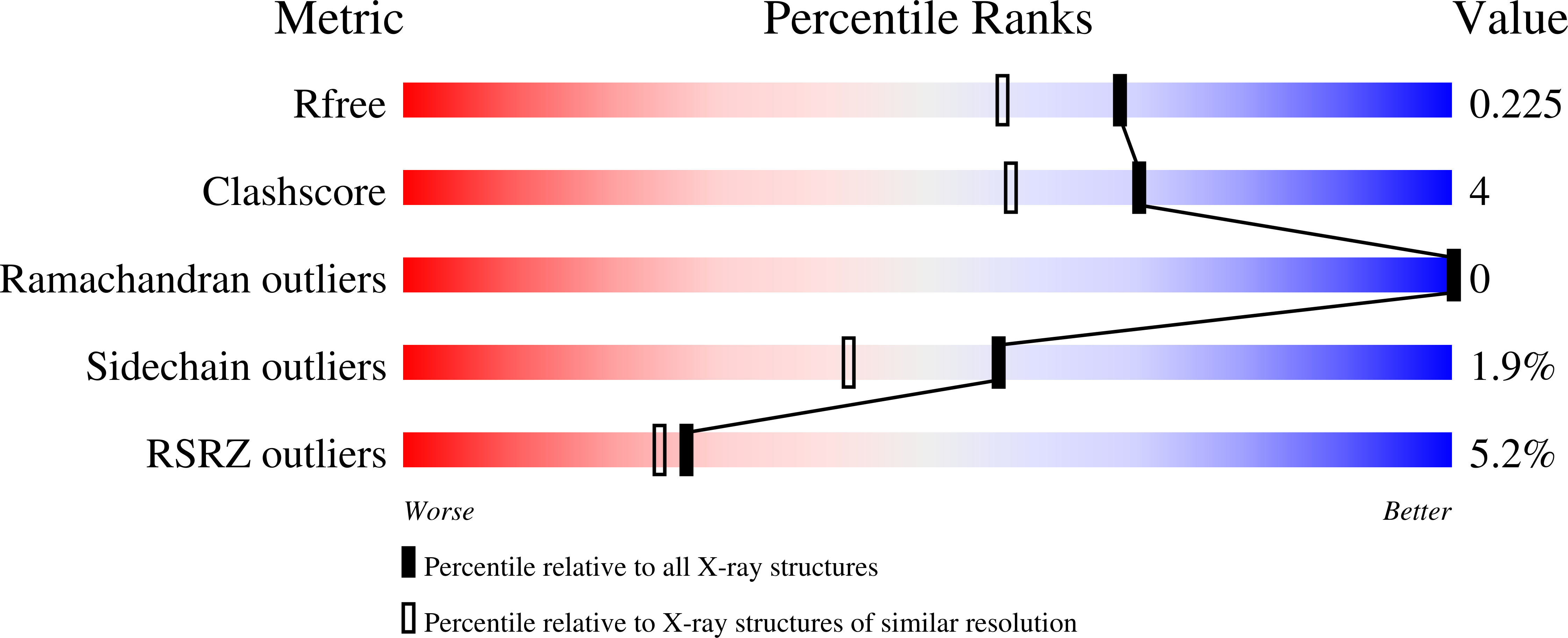
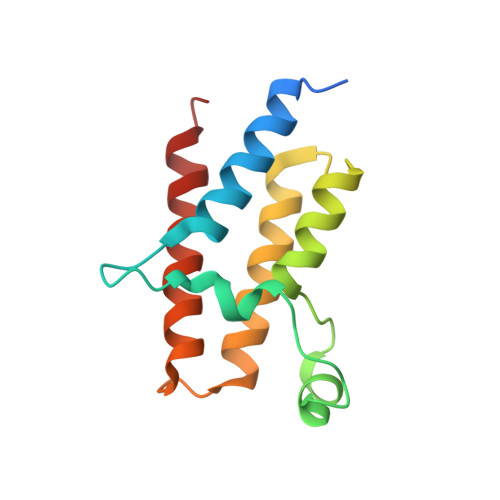
Crystal Structure of the fifth bromodomain of Human Poly-bromodomain containing protein 1 (PB1) in complex with a hydroxyphenyl-propenone ligand
Filippakopoulos, P., Picaud, S., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Knapp, S., Structural Genomics Consortium (SGC)To be published.