Recombinant production, crystallization and X-ray crystallographic structure determination of peptidyl-tRNA hydrolase from Salmonella typhimurium.
Vandavasi, V., Taylor-Creel, K., McFeeters, R.L., Coates, L., McFeeters, H.(2014) Acta Crystallogr F Struct Biol Commun 70: 872-877
- PubMed: 25005080
- DOI: https://doi.org/10.1107/S2053230X14009893
- Primary Citation of Related Structures:
4P7B - PubMed Abstract:
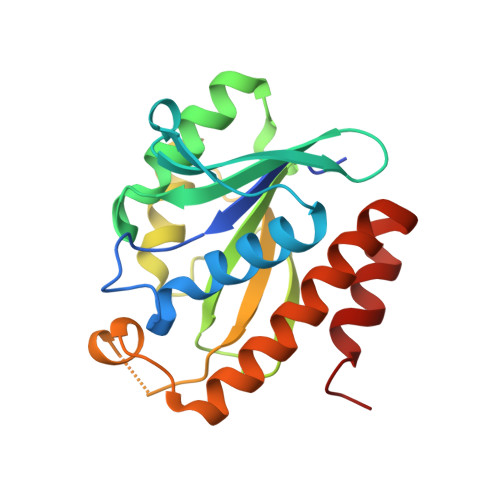
Peptidyl-tRNA hydrolase (Pth; EC 3.1.1.29) from the pathogenic bacterium Salmonella typhimurium has been cloned, expressed in Escherichia coli and crystallized for X-ray analysis. Crystals were grown using hanging-drop vapor diffusion against a reservoir solution consisting of 0.03 M citric acid, 0.05 M bis-tris propane, 1% glycerol, 3% sucrose, 25% PEG 6000 pH 7.6. Crystals were used to obtain the three-dimensional structure of the native protein at 1.6 Å resolution. The structure was determined by molecular replacement of the crystallographic data processed in space group P2₁2₁2₁ with unit-cell parameters a=62.1, b=64.9, c=110.5 Å, α=β=γ=90°. The asymmetric unit of the crystallographic lattice was composed of two copies of the enzyme molecule with a 51% solvent fraction, corresponding to a Matthews coefficient of 2.02 Å3 Da(-1). The structural coordinates reported serve as a foundation for computational and structure-guided efforts towards novel small-molecule Pth1 inhibitors and potential antibacterial development.
Organizational Affiliation:
Biology and Soft Matter Division, Oak Ridge National Laboratory, PO Box 2008, Oak Ridge, TN 37831, USA.