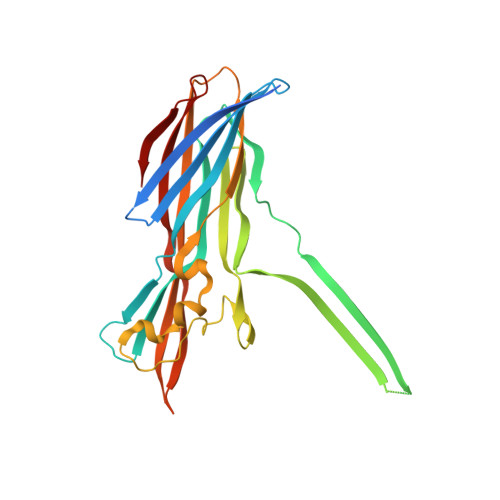
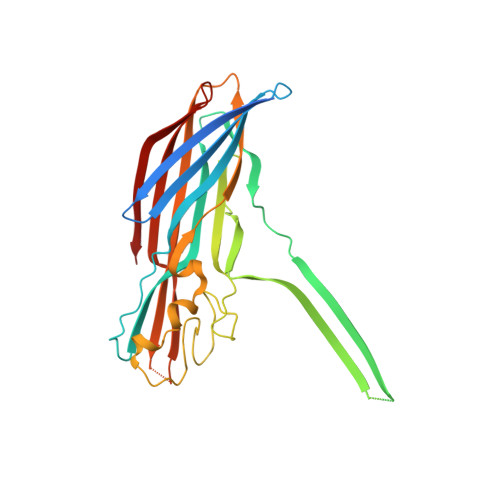
Molecular basis of transmembrane beta-barrel formation of staphylococcal pore-forming toxins.
Yamashita, D., Sugawara, T., Takeshita, M., Kaneko, J., Kamio, Y., Tanaka, I., Tanaka, Y., Yao, M.(2014) Nat Commun 5: 4897-4897
- PubMed: 25263813
- DOI: https://doi.org/10.1038/ncomms5897
- Primary Citation of Related Structures:
4P1X, 4P1Y - PubMed Abstract:
Pathogenic bacteria secrete pore-forming toxins (PFTs) to attack target cells. PFTs are expressed as water-soluble monomeric proteins, which oligomerize into nonlytic prepore intermediates on the target cell membrane before forming membrane-spanning pores. Despite a wealth of biochemical data, the lack of high-resolution prepore structural information has hampered understanding of the β-barrel formation process. Here, we report crystal structures of staphylococcal γ-haemolysin and leucocidin prepores. The structures reveal a disordered bottom half of the β-barrel corresponding to the transmembrane region, and a rigid upper extramembrane half. Spectroscopic analysis of fluorescently labelled mutants confirmed that the prepore is distinct from the pore within the transmembrane region. Mutational analysis also indicates a pivotal role for the glycine residue located at the lipid-solvent interface as a 'joint' between the two halves of the β-barrel. These observations suggest a two-step transmembrane β-barrel pore formation mechanism in which the upper extramembrane and bottom transmembrane regions are formed independently.
Organizational Affiliation:
Graduate School of Life Sciences, Hokkaido University, Sapporo 060-0810, Japan.