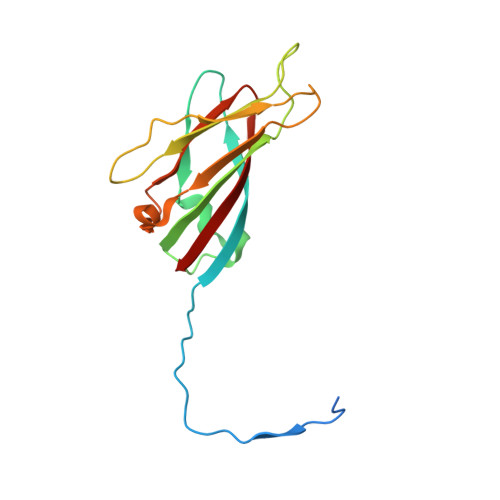
Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Larson, S.B., Day, J.S., McPherson, A.(2014) Acta Crystallogr D Biol Crystallogr 70: 2316-2330
- PubMed: 25195746
- DOI: https://doi.org/10.1107/S1399004714013789
- Primary Citation of Related Structures:
4NIA, 4OQ8, 4OQ9 - PubMed Abstract:
Satellite tobacco mosaic virus (STMV) is among the smallest viruses, having 60 identical subunits arranged with T = 1 icosahedral symmetry. Its crystal structure was solved at 290 K and was refined using, in part, crystals grown in microgravity. Electron-density maps revealed nearly 57% of the genomic ssRNA. Using six flash-cooled crystals, diffraction data were recorded to 1.4 Å resolution and independent refinements of the STMV model were carried out versus the previous 1.8 Å resolution data representing merged data from 21 crystals (271,689 unique reflections), data consisting of corresponding reflections to 1.8 Å resolution from the cooled crystals and 1.4 Å resolution data from the cooled crystals comprised of 570,721 unique reflections. Models were independently refined with full NCS constraints using the program CNS and in restrained mode using the programs CNS, REFMAC5 and SHELX-97, with the latter two procedures including anisotropic temperature factors. Significant additional structural detail emerged from the analyses, including a unique cation and anion arrangement on fivefold axes and a precise assessment of icosahedral symmetry exactness in the crystal lattice. STMV represents the highest resolution native virus structure currently known by a substantial margin, and it permits the evaluation of a precise atomic model of a spherical virus at near-atomic resolution for the first time.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, CA 92697-3900, USA.