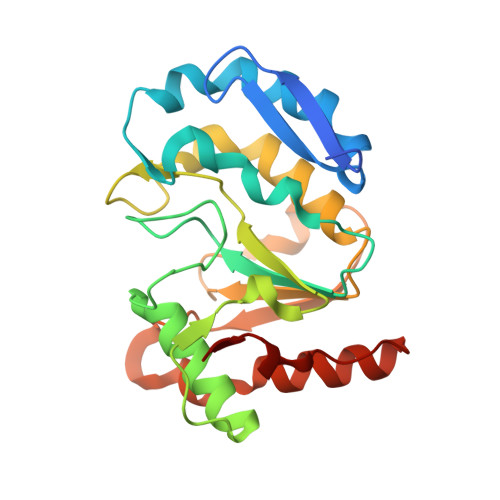
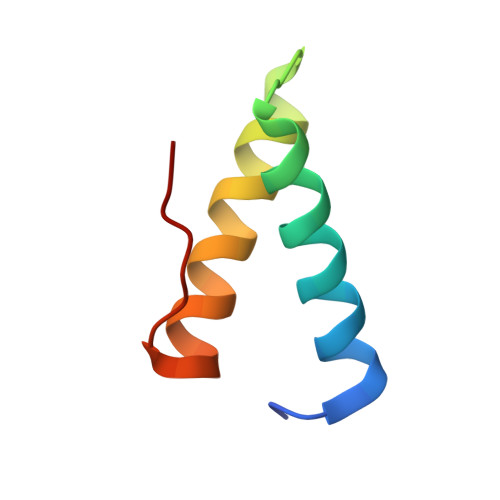
Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit d4 in complex with the a20 N-terminal domain.
Contesto-Richefeu, C., Tarbouriech, N., Brazzolotto, X., Betzi, S., Morelli, X., Burmeister, W.P., Iseni, F.(2014) PLoS Pathog 10: e1003978-e1003978
- PubMed: 24603707
- DOI: https://doi.org/10.1371/journal.ppat.1003978
- Primary Citation of Related Structures:
4OD8, 4ODA - PubMed Abstract:
Vaccinia virus polymerase holoenzyme is composed of the DNA polymerase E9, the uracil-DNA glycosylase D4 and A20, a protein with no known enzymatic activity. The D4/A20 heterodimer is the DNA polymerase co-factor whose function is essential for processive DNA synthesis. Genetic and biochemical data have established that residues located in the N-terminus of A20 are critical for binding to D4. However, no information regarding the residues of D4 involved in A20 binding is yet available. We expressed and purified the complex formed by D4 and the first 50 amino acids of A20 (D4/A20₁₋₅₀). We showed that whereas D4 forms homodimers in solution when expressed alone, D4/A20₁₋₅₀ clearly behaves as a heterodimer. The crystal structure of D4/A20₁₋₅₀ solved at 1.85 Å resolution reveals that the D4/A20 interface (including residues 167 to 180 and 191 to 206 of D4) partially overlaps the previously described D4/D4 dimer interface. A20₁₋₅₀ binding to D4 is mediated by an α-helical domain with important leucine residues located at the very N-terminal end of A20 and a second stretch of residues containing Trp43 involved in stacking interactions with Arg167 and Pro173 of D4. Point mutations of the latter residues disturb D4/A20₁₋₅₀ formation and reduce significantly thermal stability of the complex. Interestingly, small molecule docking with anti-poxvirus inhibitors selected to interfere with D4/A20 binding could reproduce several key features of the D4/A20₁₋₅₀ interaction. Finally, we propose a model of D4/A20₁₋₅₀ in complex with DNA and discuss a number of mutants described in the literature, which affect DNA synthesis. Overall, our data give new insights into the assembly of the poxvirus DNA polymerase cofactor and may be useful for the design and rational improvement of antivirals targeting the D4/A20 interface.
Organizational Affiliation:
Unité de Virologie, Institut de Recherche Biomédicale des Armées, Brétigny-sur-Orge, France.