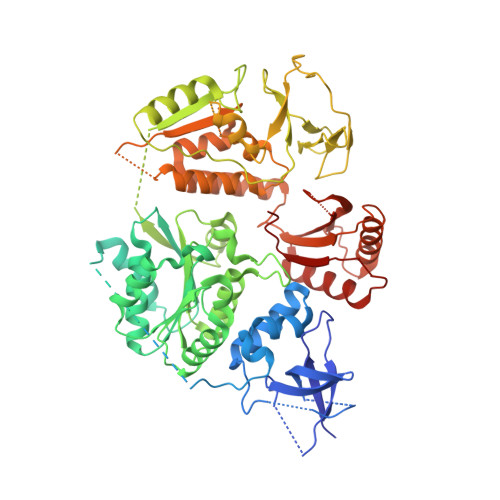
Structural mechanisms of PriA-mediated DNA replication restart.
Bhattacharyya, B., George, N.P., Thurmes, T.M., Zhou, R., Jani, N., Wessel, S.R., Sandler, S.J., Ha, T., Keck, J.L.(2014) Proc Natl Acad Sci U S A 111: 1373-1378
- PubMed: 24379377
- DOI: https://doi.org/10.1073/pnas.1318001111
- Primary Citation of Related Structures:
4NL4, 4NL8 - PubMed Abstract:
Collisions between cellular DNA replication machinery (replisomes) and damaged DNA or immovable protein complexes can dissociate replisomes before the completion of replication. This potentially lethal problem is resolved by cellular "replication restart" reactions that recognize the structures of prematurely abandoned replication forks and mediate replisomal reloading. In bacteria, this essential activity is orchestrated by the PriA DNA helicase, which identifies replication forks via structure-specific DNA binding and interactions with fork-associated ssDNA-binding proteins (SSBs). However, the mechanisms by which PriA binds replication fork DNA and coordinates subsequent replication restart reactions have remained unclear due to the dearth of high-resolution structural information available for the protein. Here, we describe the crystal structures of full-length PriA and PriA bound to SSB. The structures reveal a modular arrangement for PriA in which several DNA-binding domains surround its helicase core in a manner that appears to be poised for binding to branched replication fork DNA structures while simultaneously allowing complex formation with SSB. PriA interaction with SSB is shown to modulate SSB/DNA complexes in a manner that exposes a potential replication initiation site. From these observations, a model emerges to explain how PriA links recognition of diverse replication forks to replication restart.
Organizational Affiliation:
Department of Biomolecular Chemistry, School of Medicine and Public Health, University of Wisconsin, Madison, WI 53706.