The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Hong, J., Feng, H., Wang, F., Ranjan, A., Chen, J., Jiang, J., Ghirlando, R., Xiao, T.S., Wu, C., Bai, Y.(2014) Mol Cell 53: 498-505
- PubMed: 24507717
- DOI: https://doi.org/10.1016/j.molcel.2014.01.010
- Primary Citation of Related Structures:
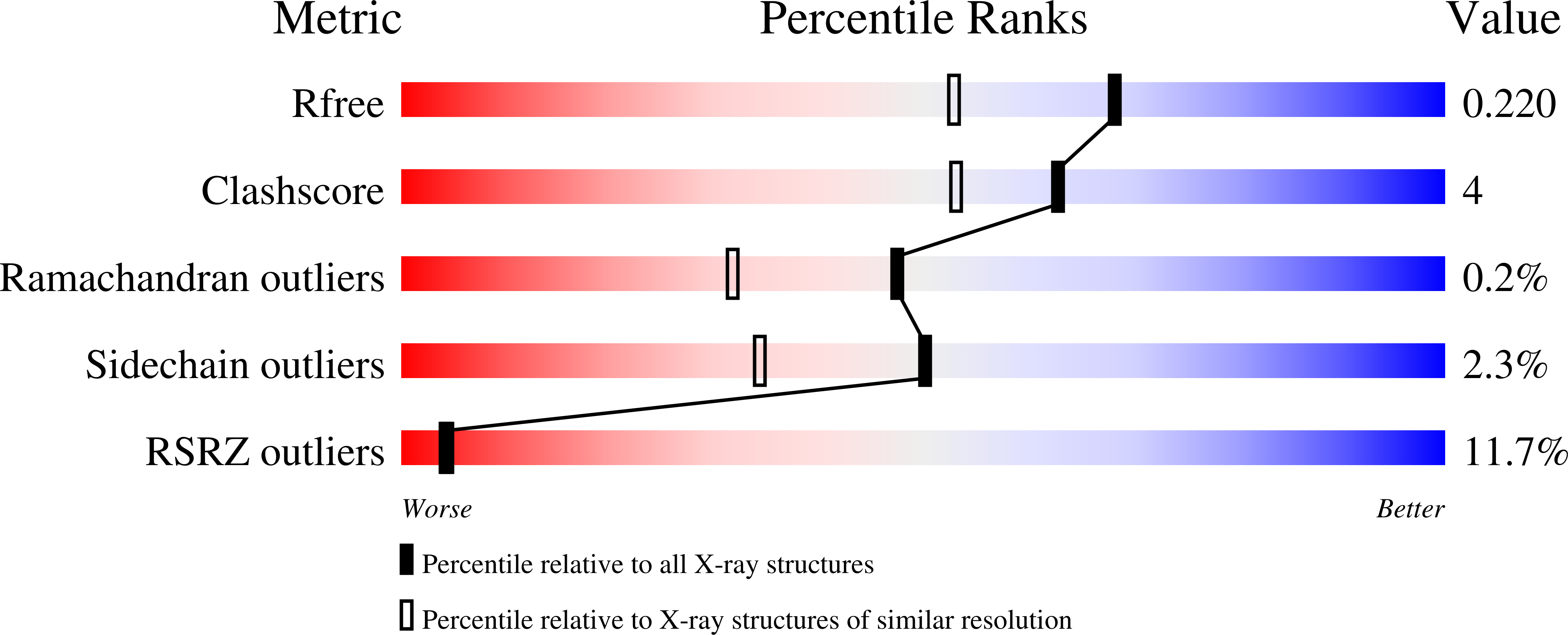
4M6B - PubMed Abstract:
Histone variant H2A.Z-containing nucleosomes exist at most eukaryotic promoters and play important roles in gene transcription and genome stability. The multisubunit nucleosome-remodeling enzyme complex SWR1, conserved from yeast to mammals, catalyzes the ATP-dependent replacement of histone H2A in canonical nucleosomes with H2A.Z. How SWR1 catalyzes the replacement reaction is largely unknown. Here, we determined the crystal structure of the N-terminal region (599-627) of the catalytic subunit Swr1, termed Swr1-Z domain, in complex with the H2A.Z-H2B dimer at 1.78 Å resolution. The Swr1-Z domain forms a 310 helix and an irregular chain. A conserved LxxLF motif in the Swr1-Z 310 helix specifically recognizes the αC helix of H2A.Z. Our results show that the Swr1-Z domain can deliver the H2A.Z-H2B dimer to the DNA-(H3-H4)2 tetrasome to form the nucleosome by a histone chaperone mechanism.
Organizational Affiliation:
Laboratory of Biochemistry and Molecular Biology, National Cancer Institute, NIH, Bethesda, MD 20892, USA.