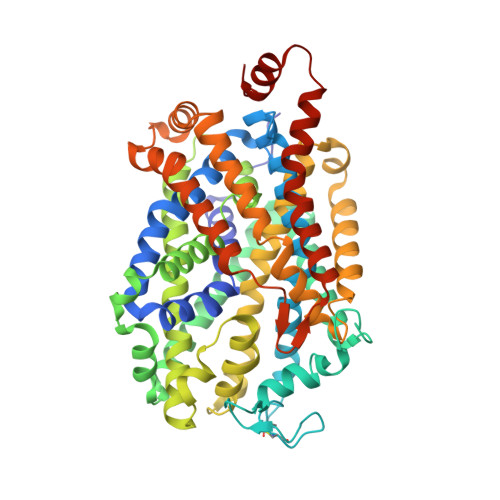
X-ray structure of dopamine transporter elucidates antidepressant mechanism.
Penmatsa, A., Wang, K.H., Gouaux, E.(2013) Nature 503: 85-90
- PubMed: 24037379
- DOI: https://doi.org/10.1038/nature12533
- Primary Citation of Related Structures:
4M48 - PubMed Abstract:
Antidepressants targeting Na(+)/Cl(-)-coupled neurotransmitter uptake define a key therapeutic strategy to treat clinical depression and neuropathic pain. However, identifying the molecular interactions that underlie the pharmacological activity of these transport inhibitors, and thus the mechanism by which the inhibitors lead to increased synaptic neurotransmitter levels, has proven elusive. Here we present the crystal structure of the Drosophila melanogaster dopamine transporter at 3.0 Å resolution bound to the tricyclic antidepressant nortriptyline. The transporter is locked in an outward-open conformation with nortriptyline wedged between transmembrane helices 1, 3, 6 and 8, blocking the transporter from binding substrate and from isomerizing to an inward-facing conformation. Although the overall structure of the dopamine transporter is similar to that of its prokaryotic relative LeuT, there are multiple distinctions, including a kink in transmembrane helix 12 halfway across the membrane bilayer, a latch-like carboxy-terminal helix that caps the cytoplasmic gate, and a cholesterol molecule wedged within a groove formed by transmembrane helices 1a, 5 and 7. Taken together, the dopamine transporter structure reveals the molecular basis for antidepressant action on sodium-coupled neurotransmitter symporters and elucidates critical elements of eukaryotic transporter structure and modulation by lipids, thus expanding our understanding of the mechanism and regulation of neurotransmitter uptake at chemical synapses.
Organizational Affiliation:
1] Vollum Institute, Oregon Health & Science University, 3181 South West Sam Jackson Park Road, Portland, Oregon 97239, USA [2].