Conformational Barrier of CheY3 and Inability of CheY4 to Bind FliM Control the Flagellar Motor Action in Vibrio cholerae
Biswas, M., Dey, S., Khamrui, S., Sen, U., Dasgupta, J.(2013) PLoS One 8: e73923-e73923
- PubMed: 24066084
- DOI: https://doi.org/10.1371/journal.pone.0073923
- Primary Citation of Related Structures:
3TO5, 4H60, 4HNQ, 4HNR, 4HNS, 4JP1, 4LX8 - PubMed Abstract:
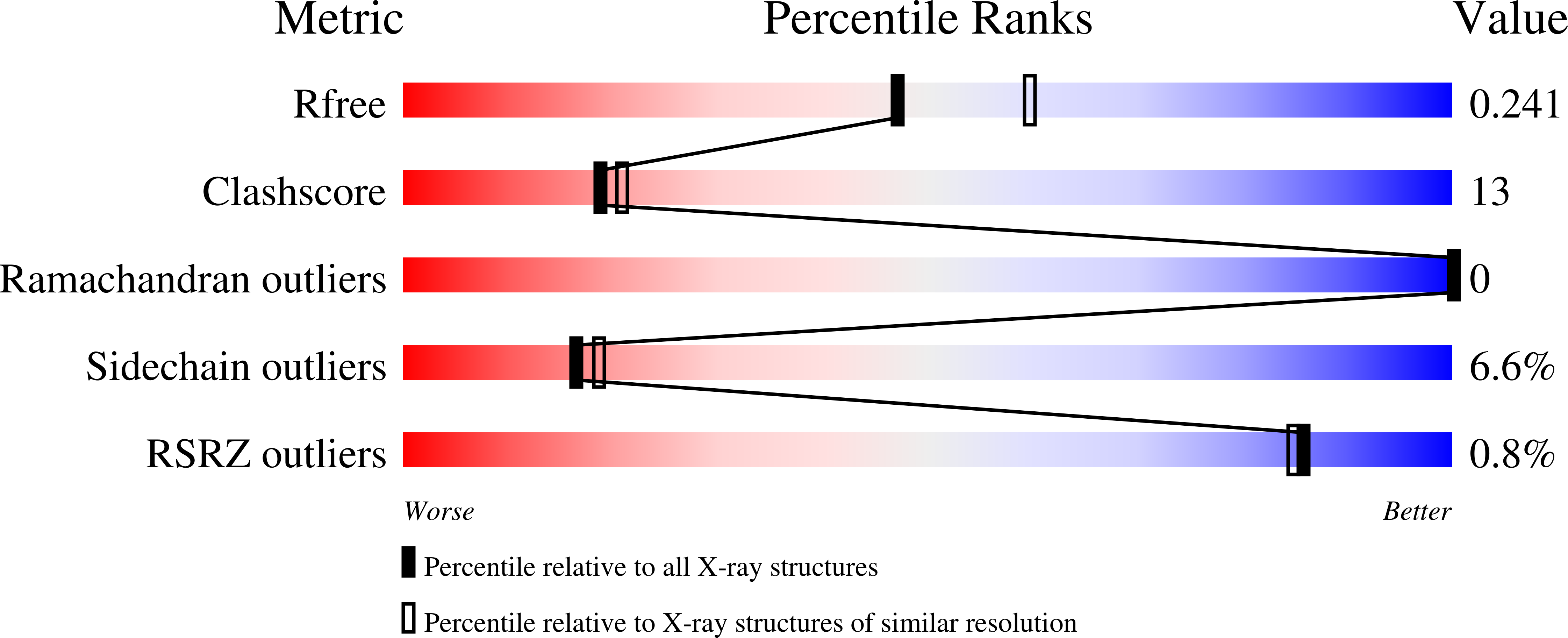
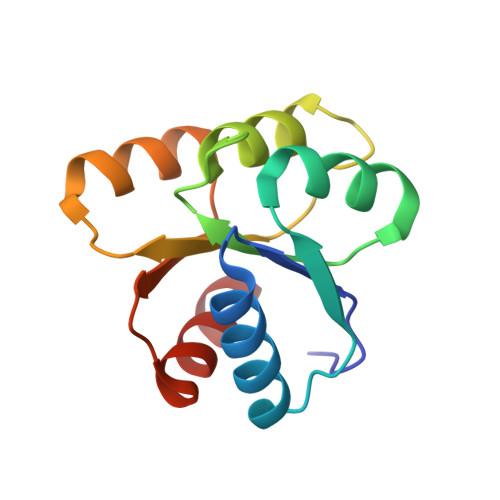
Vibrio cholerae contains multiple copies of chemotaxis response regulator (VcCheY1-VcCheY4) whose functions are elusive yet. Although previous studies suggested that only VcCheY3 directly switches the flagellar rotation, the involvement of VcCheY4 in chemotaxis could not be ruled out. None of these studies, however, focused on the structure, mechanism of activation or molecular basis of FliM binding of the VcCheYs. From the crystal structures of Ca(2+) and Mg(2+) bound VcCheY3 we proposed the presence of a conformational barrier composed of the hydrophobic packing of W61, M88 and V106 and a unique hydrogen bond between T90 and Q97 in VcCheY3. Lesser fluorescence quenching and higher Km value of VcCheY3, compared to its mutants VcCheY3-Q97A and VcCheY3-Q97A/E100A supported our proposition. Furthermore, aforesaid biochemical data, in conjunction with the structure of VcCheY3-Q97A, indicated that the coupling of T90 and Q97 restricts the movement of T90 toward the active site reducing the stabilization of the bound phosphate and effectively promoting autodephosphorylation of VcCheY3. The structure of BeF3(-) activated VcCheY3 insisted us to argue that elevated temperature and/or adequacy of phosphate pool might break the barrier of the free-state VcCheY3 and the conformational changes, required for FliM binding, occur upon phosphorylation. Structure of VcCheY4 has been solved in the free and sulfated states. VcCheY4(sulf), containing a bound sulfate at the active site, appears to be more compact and stable with a longer α4 helix, shorter β4α4 loop and hydrogen bond between T82 and the sulfate compared to VcCheY4(free). While pull down assay of VcCheYs with VcFliMNM showed that only activated VcCheY3 can interact with VcFliMNM and VcCheY4 cannot, a knowledge based docking explained the molecular mechanism of the interactions between VcCheY3 and VcFliM and identified the limitations of VcCheY4 to interact with VcFliM even in its phosphorylated state.
Organizational Affiliation:
Department of Biotechnology, St. Xavier's College, Kolkata, India.