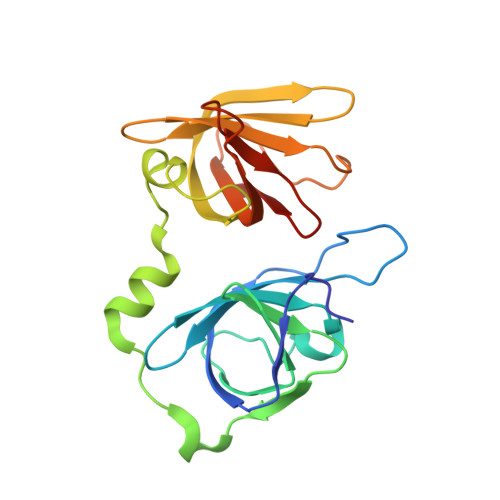
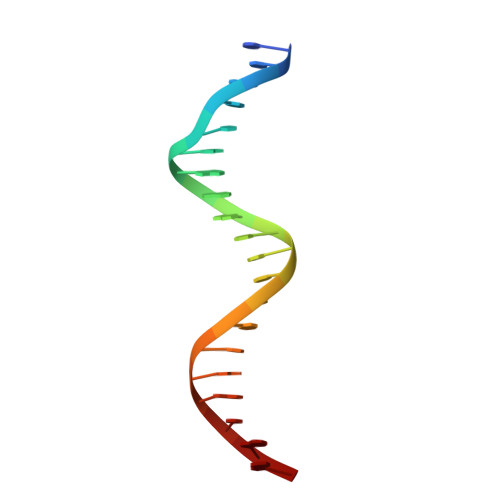
Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling.
Li, H., Wang, J., Wang, J., Cao, L.S., Wang, Z.X., Wu, J.W.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 21-29
- PubMed: 24419611
- DOI: https://doi.org/10.1107/S2053230X1303135X
- Primary Citation of Related Structures:
4LNQ - PubMed Abstract:
The HIN-200 family of proteins play significant roles in inflammation-related processes. Among them, AIM2 (absent in melanoma 2) and IFI16 (γ-interferon-inducible protein 16) recognize double-stranded DNA to initiate inflammatory responses. In contrast, p202, a mouse interferon-inducible protein containing two HIN domains (HINa and HINb), has been reported to inhibit Aim2-mediated inflammatory signalling in mouse. To understand the inhibitory mechanism, the crystal structure of the p202 HINa domain in complex with a 20 bp DNA was determined, in which p202 HINa nonspecifically recognizes both strands of DNA through electrostatic attraction. The p202 HINa domain binds DNA more tightly than does AIM2 HIN, and the DNA-binding mode of p202 HINa is different from that of the AIM2 HIN and IFI16 HINb domains. These results, together with the reported data on p202 HINb, lead to an interaction model for full-length p202 and dsDNA which provides a conceivable mechanism for the negative regulation of Aim2 inflammasome activation by p202.
Organizational Affiliation:
MOE Key Laboratory of Protein Science and Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, People's Republic of China.