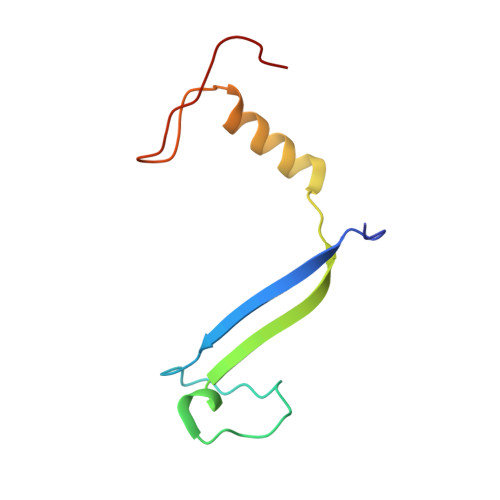
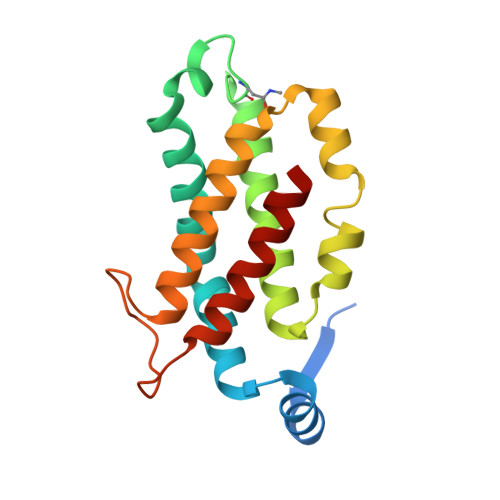
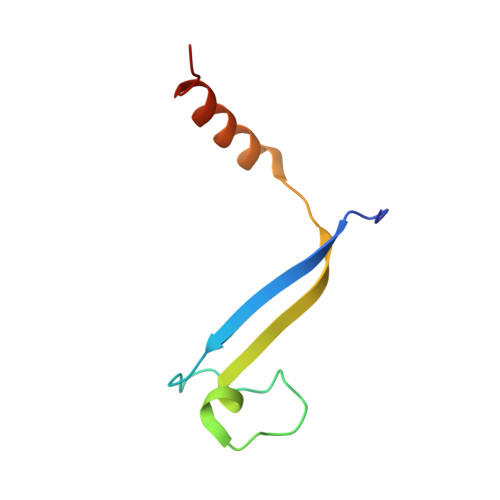
Single-residue insertion switches the quaternary structure and exciton states of cryptophyte light-harvesting proteins.
Harrop, S.J., Wilk, K.E., Dinshaw, R., Collini, E., Mirkovic, T., Teng, C.Y., Oblinsky, D.G., Green, B.R., Hoef-Emden, K., Hiller, R.G., Scholes, G.D., Curmi, P.M.(2014) Proc Natl Acad Sci U S A 111: E2666-E2675
- PubMed: 24979784
- DOI: https://doi.org/10.1073/pnas.1402538111
- Primary Citation of Related Structures:
4LM6, 4LMS, 4LMX - PubMed Abstract:
Observation of coherent oscillations in the 2D electronic spectra (2D ES) of photosynthetic proteins has led researchers to ask whether nontrivial quantum phenomena are biologically significant. Coherent oscillations have been reported for the soluble light-harvesting phycobiliprotein (PBP) antenna isolated from cryptophyte algae. To probe the link between spectral properties and protein structure, we determined crystal structures of three PBP light-harvesting complexes isolated from different species. Each PBP is a dimer of αβ subunits in which the structure of the αβ monomer is conserved. However, we discovered two dramatically distinct quaternary conformations, one of which is specific to the genus Hemiselmis. Because of steric effects emerging from the insertion of a single amino acid, the two αβ monomers are rotated by ∼73° to an "open" configuration in contrast to the "closed" configuration of other cryptophyte PBPs. This structural change is significant for the light-harvesting function because it disrupts the strong excitonic coupling between two central chromophores in the closed form. The 2D ES show marked cross-peak oscillations assigned to electronic and vibrational coherences in the closed-form PC645. However, such features appear to be reduced, or perhaps absent, in the open structures. Thus cryptophytes have evolved a structural switch controlled by an amino acid insertion to modulate excitonic interactions and therefore the mechanisms used for light harvesting.
Organizational Affiliation:
School of Physics, The University of New South Wales, Sydney, NSW 2052, Australia;