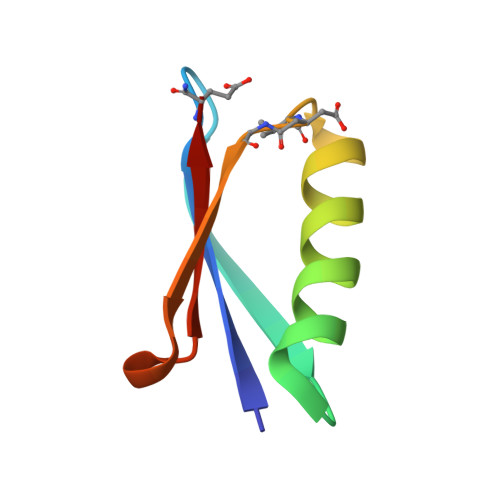
Protein-like Tertiary Folding Behavior from Heterogeneous Backbones.
Reinert, Z.E., Lengyel, G.A., Horne, W.S.(2013) J Am Chem Soc 135: 12528-12531
- PubMed: 23937097
- DOI: https://doi.org/10.1021/ja405422v
- Primary Citation of Related Structures:
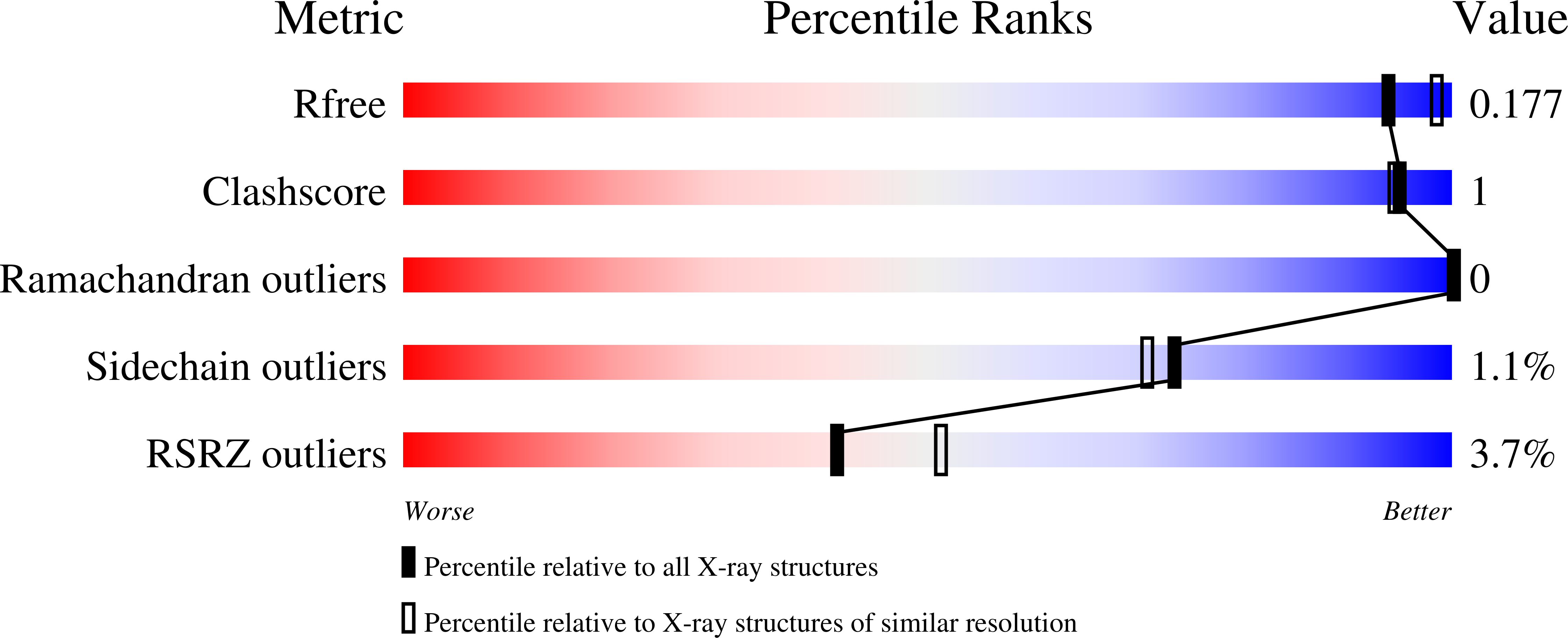
4KGR, 4KGS, 4KGT - PubMed Abstract:
Because proteins play vital roles in life, much effort has been invested in their mimicry by synthetic agents. One approach is to design unnatural backbone oligomers ("foldamers") that fold like natural peptides. Despite success in secondary structure mimicry by such species, protein-like tertiary folds remain elusive. A fundamental challenge underlying this task is the design of a sequence of side chains that will specify a complex tertiary folding pattern on an unnatural backbone. We report here a sequence-based approach to convert a natural protein with a compact tertiary fold to an analogue with a backbone composed of ~20% unnatural building blocks but folding behavior similar to that of the parent protein.
Organizational Affiliation:
Department of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, USA.