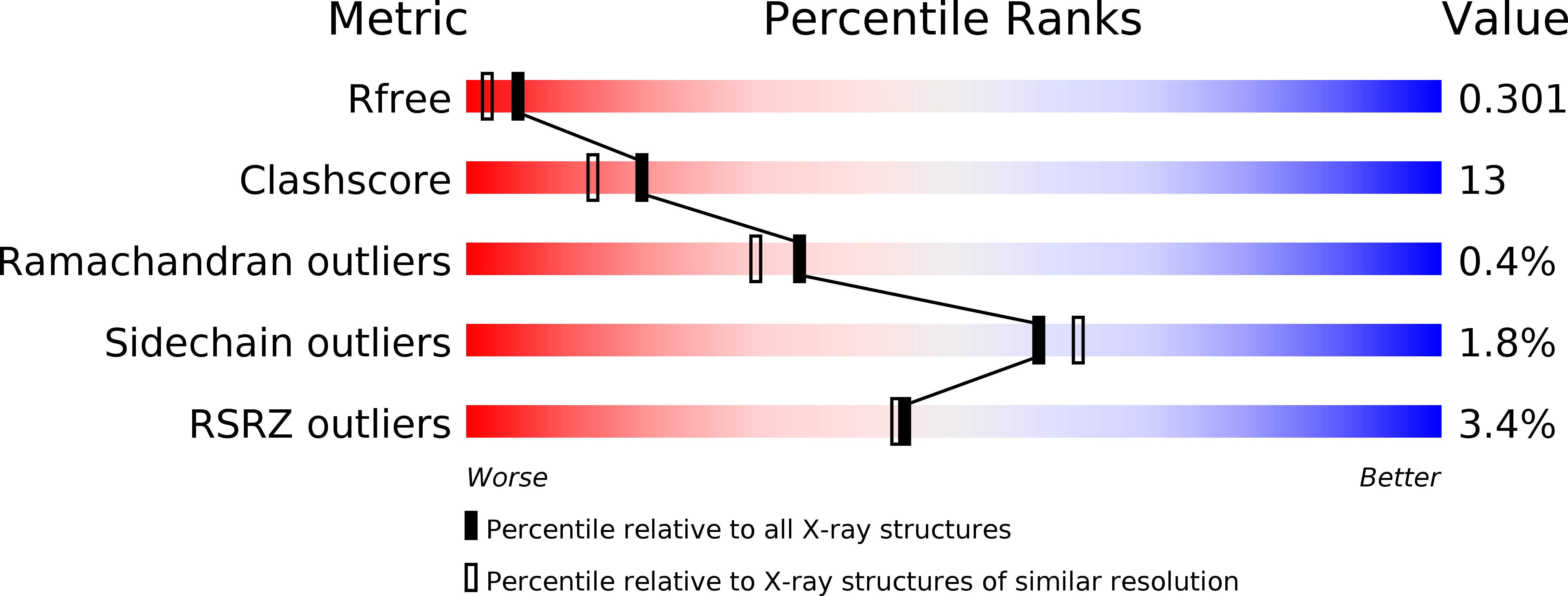
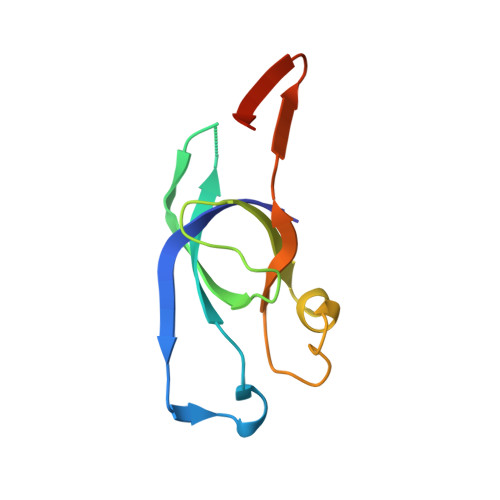
Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species.
Sutter, M., Wilson, S.C., Deutsch, S., Kerfeld, C.A.(2013) Photosynth Res 118: 9-16
- PubMed: 23949415
- DOI: https://doi.org/10.1007/s11120-013-9909-z
- Primary Citation of Related Structures:
4JVZ, 4JW0 - PubMed Abstract:
Cyanobacteria have evolved a unique carbon fixation organelle known as the carboxysome that compartmentalizes the enzymes RuBisCO and carbonic anhydrase. This effectively increases the local CO2 concentration at the active site of RuBisCO and decreases its relatively unproductive side reaction with oxygen. Carboxysomes consist of a protein shell composed of hexameric and pentameric proteins arranged in icosahedral symmetry. Facets composed of hexameric proteins are connected at the vertices by pentameric proteins. Structurally homologous pentamers and hexamers are also found in heterotrophic bacteria where they form architecturally related microcompartments such as the Eut and Pdu organelles for the metabolism of ethanolamine and propanediol, respectively. Here we describe two new high-resolution structures of the pentameric shell protein CcmL from the cyanobacteria Thermosynechococcus elongatus and Gloeobacter violaceus and provide detailed analysis of their characteristics and comparison with related shell proteins.
Organizational Affiliation:
United States Department of Energy - Joint Genome Institute, Walnut Creek, CA, 94598, USA.