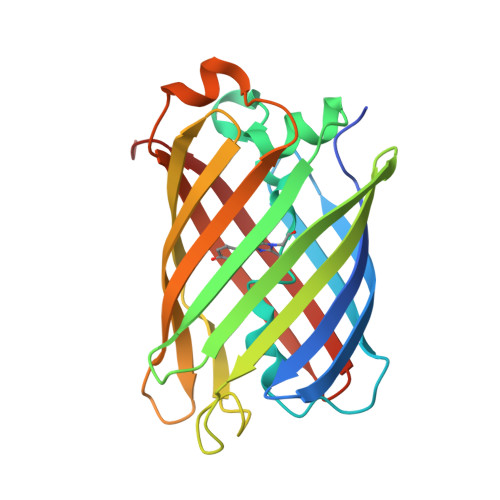
Structure of the red fluorescent protein from a lancelet (Branchiostoma lanceolatum): a novel GYG chromophore covalently bound to a nearby tyrosine.
Pletnev, V.Z., Pletneva, N.V., Lukyanov, K.A., Souslova, E.A., Fradkov, A.F., Chudakov, D.M., Chepurnykh, T., Yampolsky, I.V., Wlodawer, A., Dauter, Z., Pletnev, S.(2013) Acta Crystallogr D Biol Crystallogr 69: 1850-1860
- PubMed: 23999308
- DOI: https://doi.org/10.1107/S0907444913015424
- Primary Citation of Related Structures:
4HVF, 4JEO, 4JF9, 4JGE - PubMed Abstract:
A key property of proteins of the green fluorescent protein (GFP) family is their ability to form a chromophore group by post-translational modifications of internal amino acids, e.g. Ser65-Tyr66-Gly67 in GFP from the jellyfish Aequorea victoria (Cnidaria). Numerous structural studies have demonstrated that the green GFP-like chromophore represents the `core' structure, which can be extended in red-shifted proteins owing to modifications of the protein backbone at the first chromophore-forming position. Here, the three-dimensional structures of green laGFP (λex/λem = 502/511 nm) and red laRFP (λex/λem ≃ 521/592 nm), which are fluorescent proteins (FPs) from the lancelet Branchiostoma lanceolatum (Chordata), were determined together with the structure of a red variant laRFP-ΔS83 (deletion of Ser83) with improved folding. Lancelet FPs are evolutionarily distant and share only ∼20% sequence identity with cnidarian FPs, which have been extensively characterized and widely used as genetically encoded probes. The structure of red-emitting laRFP revealed three exceptional features that have not been observed in wild-type fluorescent proteins from Cnidaria reported to date: (i) an unusual chromophore-forming sequence Gly58-Tyr59-Gly60, (ii) the presence of Gln211 at the position of the conserved catalytic Glu (Glu222 in Aequorea GFP), which proved to be crucial for chromophore formation, and (iii) the absence of modifications typical of known red chromophores and the presence of an extremely unusual covalent bond between the Tyr59 C(β) atom and the hydroxyl of the proximal Tyr62. The impact of this covalent bond on the red emission and the large Stokes shift (∼70 nm) of laRFP was verified by extensive structure-based site-directed mutagenesis.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russian Federation.