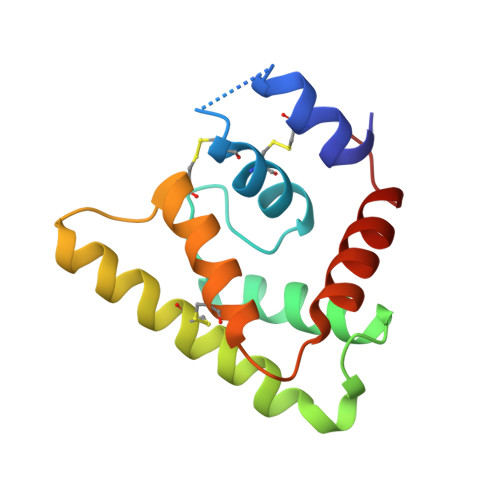
Novel family of insect salivary inhibitors blocks contact pathway activation by binding to polyphosphate, heparin, and dextran sulfate.
Alvarenga, P.H., Xu, X., Oliveira, F., Chagas, A.C., Nascimento, C.R., Francischetti, I.M., Juliano, M.A., Juliano, L., Scharfstein, J., Valenzuela, J.G., Ribeiro, J.M., Andersen, J.F.(2013) Arterioscler Thromb Vasc Biol 33: 2759-2770
- PubMed: 24092749
- DOI: https://doi.org/10.1161/ATVBAHA.113.302482
- Primary Citation of Related Structures:
4JD9 - PubMed Abstract:
Polyphosphate and heparin are anionic polymers released by activated mast cells and platelets that are known to stimulate the contact pathway of coagulation. These polymers promote both the autoactivation of factor XII and the assembly of complexes containing factor XI, prekallikrein, and high-molecular-weight kininogen. We are searching for salivary proteins from blood-feeding insects that counteract the effect of procoagulant and proinflammatory factors in the host, including elements of the contact pathway. Here, we evaluate the ability of the sand fly salivary proteins, PdSP15a and PdSP15b, to inhibit the contact pathway by disrupting binding of its components to anionic polymers. We attempt to demonstrate binding of the proteins to polyphosphate, heparin, and dextran sulfate. We also evaluate the effect of this binding on contact pathway reactions. We also set out to determine the x-ray crystal structure of PdSP15b and examine the determinants of relevant molecular interactions. Both proteins bind polyphosphate, heparin, and dextran sulfate with high affinity. Through this mechanism they inhibit the autoactivation of factor XII and factor XI, the reciprocal activation of factor XII and prekallikrein, the activation of factor XI by thrombin and factor XIIa, the cleavage of high-molecular-weight kininogen in plasma, and plasma extravasation induced by polyphosphate. The crystal structure of PdSP15b contains an amphipathic helix studded with basic side chains that forms the likely interaction surface. The results of these studies indicate that the binding of anionic polymers by salivary proteins is used by blood feeders as an antihemostatic/anti-inflammatory mechanism.
Organizational Affiliation:
From the Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Rockville, MD (P.H.A., X.X., F.O., A.C.C., I.M.B.F., J.G.V., J.M.C.R., J.F.A.); Laboratório de Bioquímica de Resposta ao Estresse, Instituto de Bioquímica Médica, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil (P.H.A.); Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular (INCT-EM), Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil (P.H.A.); Instituto de Biofísica Carlos Chagas Filho, Centro de Ciências da Saúde, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil (C.R.N., J.S.); and Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, Brazil (M.A.J., L.J.).