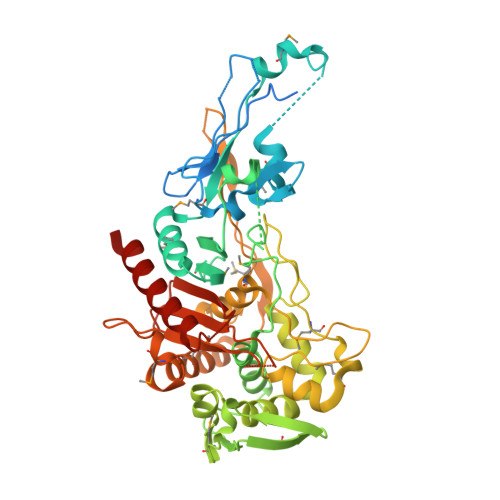
Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
Filippova, E.V., Wawrzak, Z., Minasov, G., Shuvalova, L., Kiryukhina, O., Babnigg, G., Rubin, E., Sacchettini, J., Joachimiak, A., Anderson, W.F., Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI)To be published.