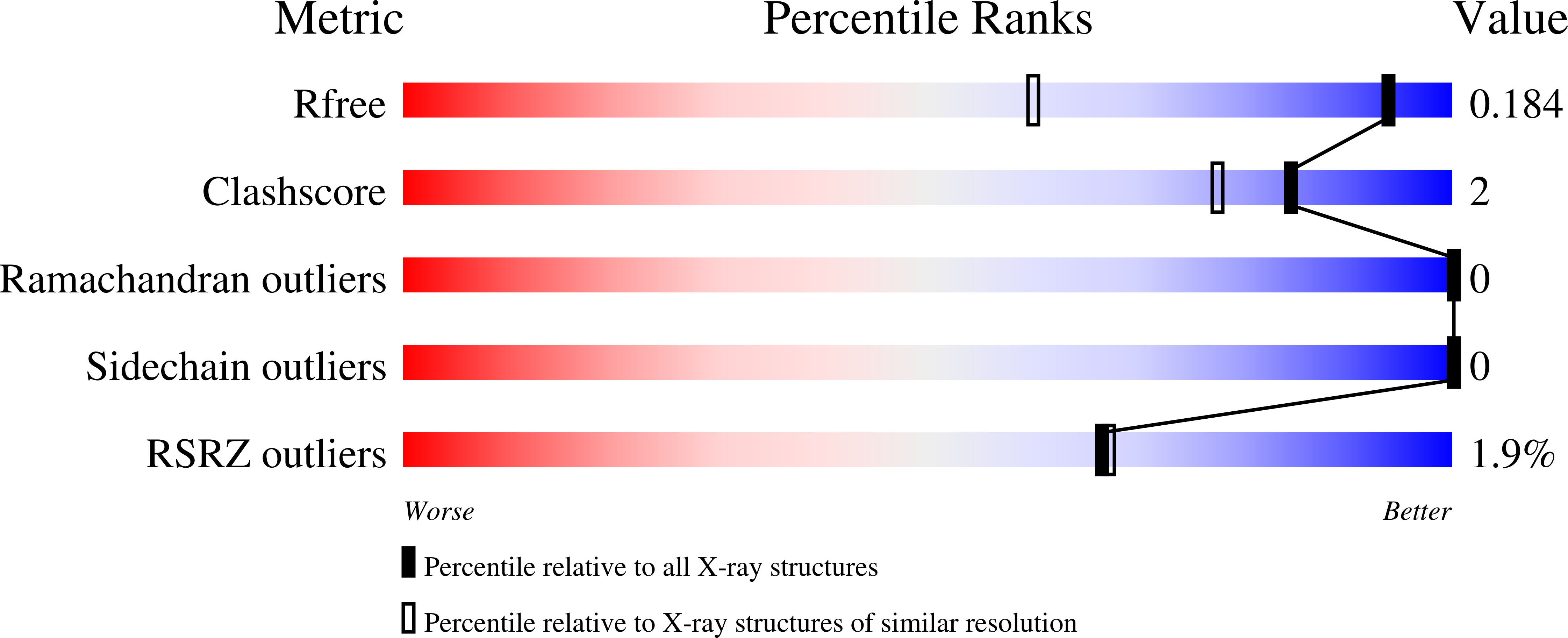
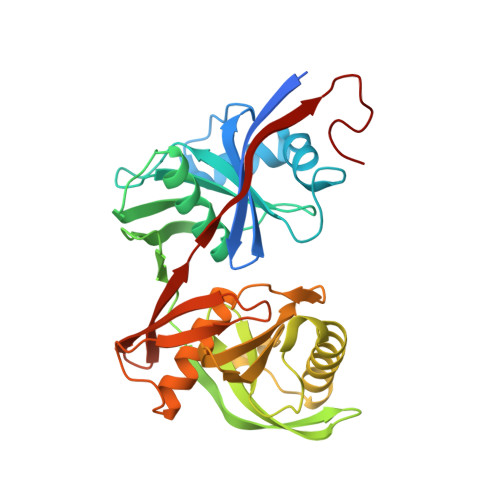
Crystal structure of pput_1285, a putative hydroxyproline epimerase from pseudomonas putida f1 (target efi-506500), open form, space group i2, bound citrate
Vetting, M.W., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hammonds, J., Stead, M., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Gerlt, J.A., Almo, S.C.To be published.