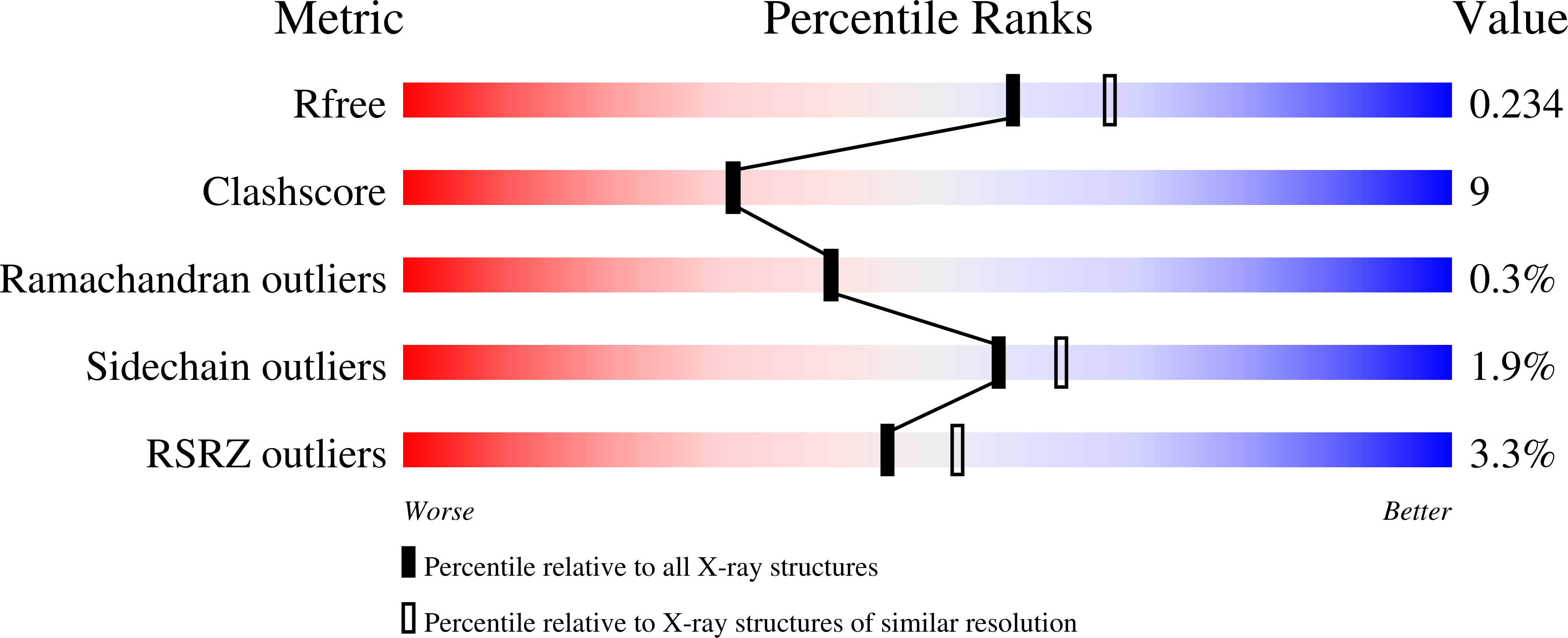
Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.
Chen, M.W., Lohkamp, B., Schnell, R., Lescar, J., Schneider, G.(2013) PLoS One 8: e66936-e66936
- PubMed: 23805286
- DOI: https://doi.org/10.1371/journal.pone.0066936
- Primary Citation of Related Structures:
4JAY, 4JB1 - PubMed Abstract:
Biosynthesis of UDP-N-acetylmuramic acid in bacteria is a committed step towards peptidoglycan production. In an NADPH- and FAD-dependent reaction, the UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) reduces UDP-N-acetylglucosamine-enolpyruvate to UDP-N-acetylmuramic acid. We determined the three-dimensional structures of the ternary complex of Pseudomonas aeruginosa MurB with FAD and NADP(+) in two crystal forms to resolutions of 2.2 and 2.1 Å, respectively, to investigate the structural basis of the first half-reaction, hydride transfer from NADPH to FAD. The nicotinamide ring of NADP(+) stacks against the si face of the isoalloxazine ring of FAD, suggesting an unusual mode of hydride transfer to flavin. Comparison with the structure of the Escherichia coli MurB complex with UDP-N-acetylglucosamine-enolpyruvate shows that both substrates share the binding site located between two lobes of the substrate-binding domain III, consistent with a ping pong mechanism with sequential substrate binding. The nicotinamide and the enolpyruvyl moieties are strikingly well-aligned upon superimposition, both positioned for hydride transfer to and from FAD. However, flexibility of the substrate channel allows the non-reactive parts of the two substrates to bind in different conformations. A potassium ion in the active site may assist in substrate orientation and binding. These structural models should help in structure-aided drug design against MurB, which is essential for cell wall biogenesis and hence bacterial survival.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden ; School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.