The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators.
Alanazi, A.M., Neidle, E.L., Momany, C.(2013) Acta Crystallogr D Biol Crystallogr 69: 1995-2007
- PubMed: 24100318
- DOI: https://doi.org/10.1107/S0907444913017320
- Primary Citation of Related Structures:
4IHS, 4IHT - PubMed Abstract:
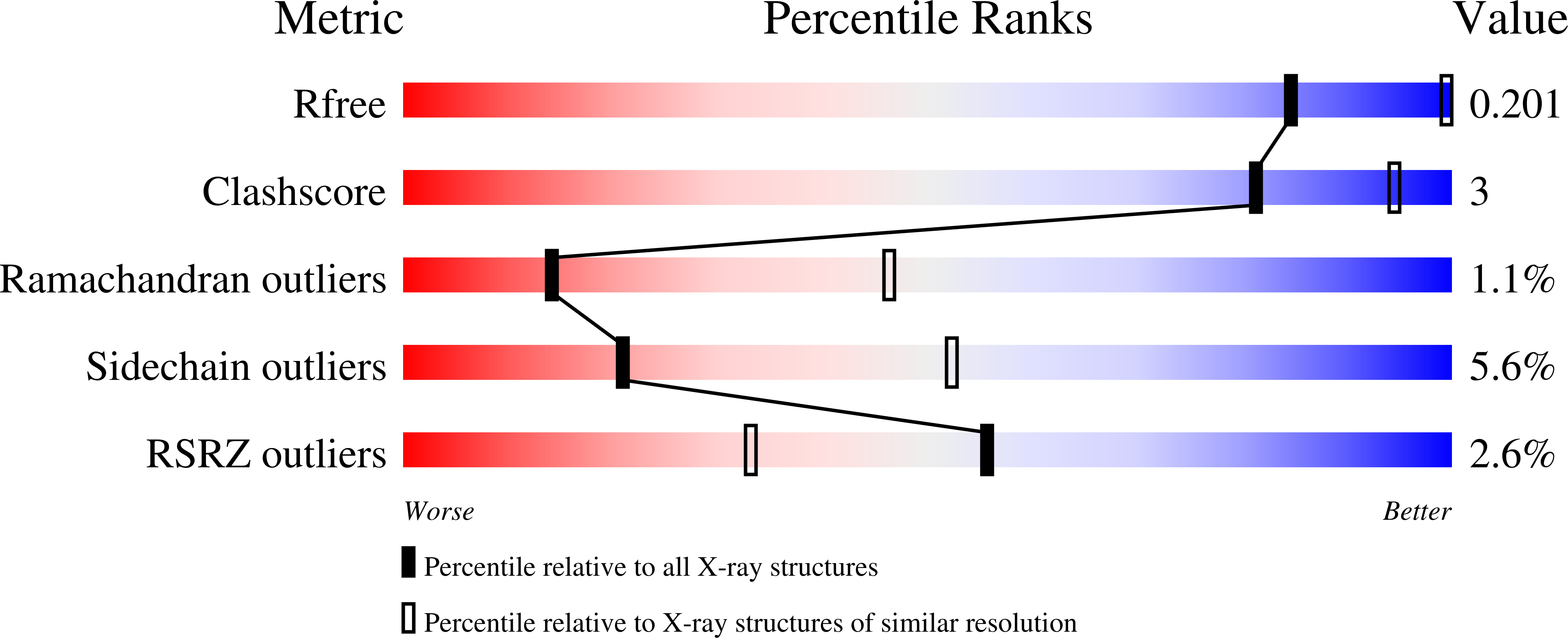
LysR-type transcriptional regulators (LTTRs) play critical roles in metabolism and constitute the largest family of bacterial regulators. To understand protein-DNA interactions, atomic structures of the DNA-binding domain and linker-helix regions of a prototypical LTTR, BenM, were determined by X-ray crystallography. BenM structures with and without bound DNA reveal a set of highly conserved amino acids that interact directly with DNA bases. At the N-terminal end of the recognition helix (α3) of a winged-helix-turn-helix DNA-binding motif, several residues create hydrophobic pockets (Pro30, Pro31 and Ser33). These pockets interact with the methyl groups of two thymines in the DNA-recognition motif and its complementary strand, T-N11-A. This motif usually includes some dyad symmetry, as exemplified by a sequence that binds two subunits of a BenM tetramer (ATAC-N7-GTAT). Gln29 forms hydrogen bonds to adenine in the first position of the recognition half-site (ATAC). Another hydrophobic pocket defined by Ala28, Pro30 and Pro31 interacts with the methyl group of thymine, complementary to the base at the third position of the half-site. Arg34 interacts with the complementary base of the 3' position. Arg53, in the wing, provides AT-tract recognition in the minor groove. For DNA recognition, LTTRs use highly conserved interactions between amino acids and nucleotide bases as well as numerous less-conserved secondary interactions.
Organizational Affiliation:
Department of Pharmaceutical and Biomedical Sciences, University of Georgia, Athens, GA 30602, USA.