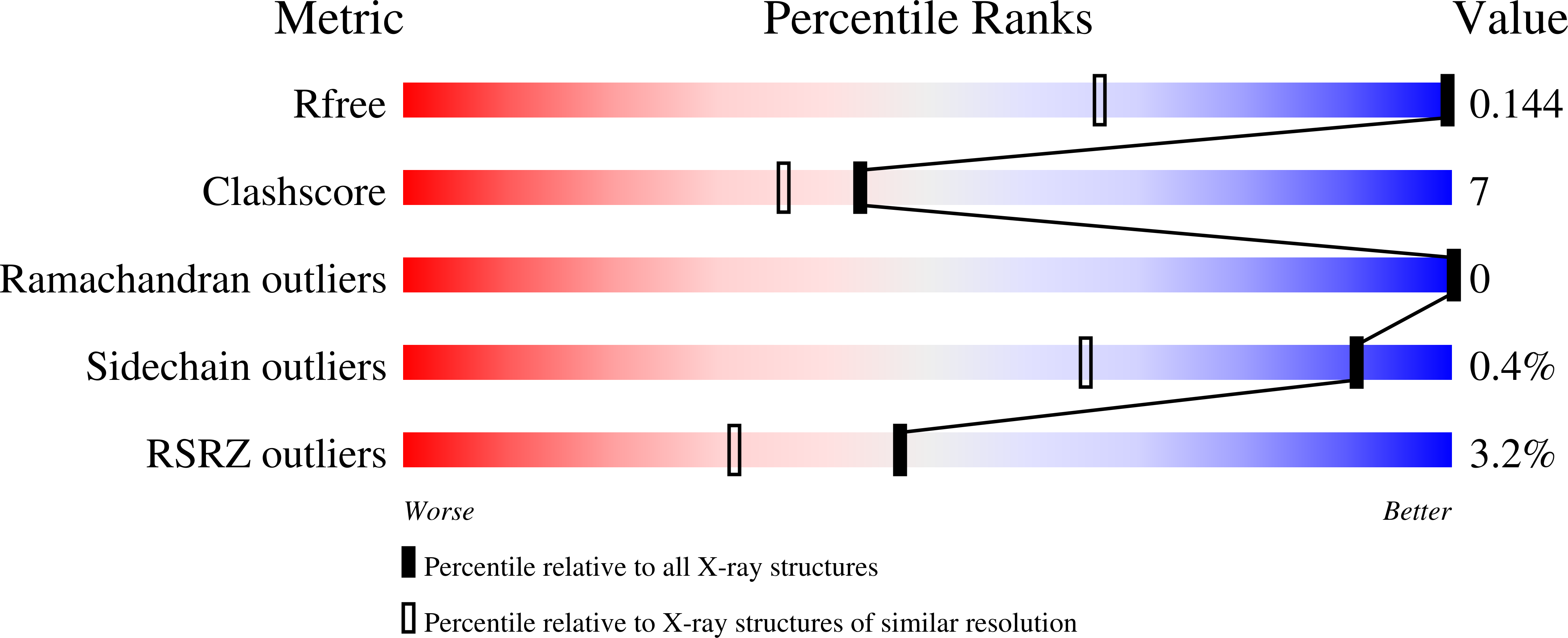
Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Cousido-Siah, A., Ruiz, F.X., Mitschler, A., Porte, S., de Lera, A.R., Martin, M.J., Manzanaro, S., de la Fuente, J.A., Terwesten, F., Betz, M., Klebe, G., Farres, J., Pares, X., Podjarny, A.(2014) Acta Crystallogr D Biol Crystallogr 70: 889-903
- PubMed: 24598757
- DOI: https://doi.org/10.1107/S1399004713033452
- Primary Citation of Related Structures:
4ICC, 4IGS - PubMed Abstract:
Aldo-keto reductases (AKRs) are mostly monomeric enzymes which fold into a highly conserved (α/β)8 barrel, while their substrate specificity and inhibitor selectivity are determined by interaction with residues located in three highly variable external loops. The closely related human enzymes aldose reductase (AR or AKR1B1) and AKR1B10 are of biomedical interest because of their involvement in secondary diabetic complications (AR) and in cancer, e.g. hepatocellular carcinoma and smoking-related lung cancer (AKR1B10). After characterization of the IC50 values of both AKRs with a series of polyhalogenated compounds, 2,2',3,3',5,5',6,6'-octafluoro-4,4'-biphenyldiol (JF0064) was identified as a lead inhibitor of both enzymes with a new scaffold (a 1,1'-biphenyl-4,4'-diol). An ultrahigh-resolution X-ray structure of the AR-NADP(+)-JF0064 complex has been determined at 0.85 Å resolution, allowing it to be observed that JF0064 interacts with the catalytic residue Tyr48 through a negatively charged hydroxyl group (i.e. the acidic phenol). The non-competitive inhibition pattern observed for JF0064 with both enzymes suggests that this acidic hydroxyl group is also present in the case of AKR1B10. Moreover, the combination of surface lysine methylation and the introduction of K125R and V301L mutations enabled the determination of the X-ray crystallographic structure of the corresponding AKR1B10-NADP(+)-JF0064 complex. Comparison of the two structures has unveiled some important hints for subsequent structure-based drug-design efforts.
Organizational Affiliation:
Department of Integrative Biology, Institut de Génétique et de Biologie Moléculaire et Cellulaire, CNRS/INSER/UdS, 1 Rue Laurent Fries, 67404 Illkirch CEDEX, France.