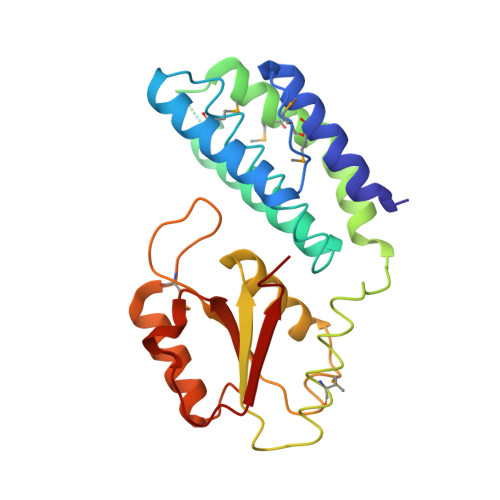
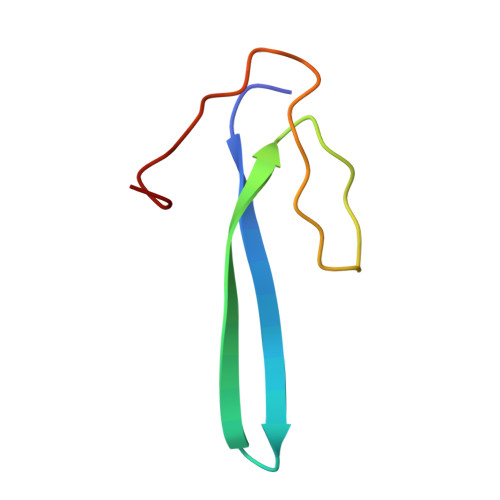
Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling.
Motz, C., Schuhmann, K.M., Kirchhofer, A., Moldt, M., Witte, G., Conzelmann, K.K., Hopfner, K.P.(2013) Science 339: 690-693
- PubMed: 23328395
- DOI: https://doi.org/10.1126/science.1230949
- Primary Citation of Related Structures:
4I1S - PubMed Abstract:
The retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) melanoma differentiation-associated protein 5 (MDA5) senses cytoplasmic viral RNA and activates antiviral innate immunity. To reveal how paramyxoviruses counteract this response, we determined the crystal structure of the MDA5 adenosine 5'-triphosphate (ATP)-hydrolysis domain in complex with the viral inhibitor V protein. The V protein unfolded the ATP-hydrolysis domain of MDA5 via a β-hairpin motif and recognized a structural motif of MDA5 that is normally buried in the conserved helicase fold. This leads to disruption of the MDA5 ATP-hydrolysis site and prevention of RNA-bound MDA5 filament formation. The structure explains why V proteins inactivate MDA5, but not RIG-I, and mutating only two amino acids in RIG-I induces robust V protein binding. Our results suggest an inhibition mechanism of RLR signalosome formation by unfolding of receptor and inhibitor.
Organizational Affiliation:
Department of Biochemistry and Gene Center, Ludwig-Maximilians-University, Munich, Germany.