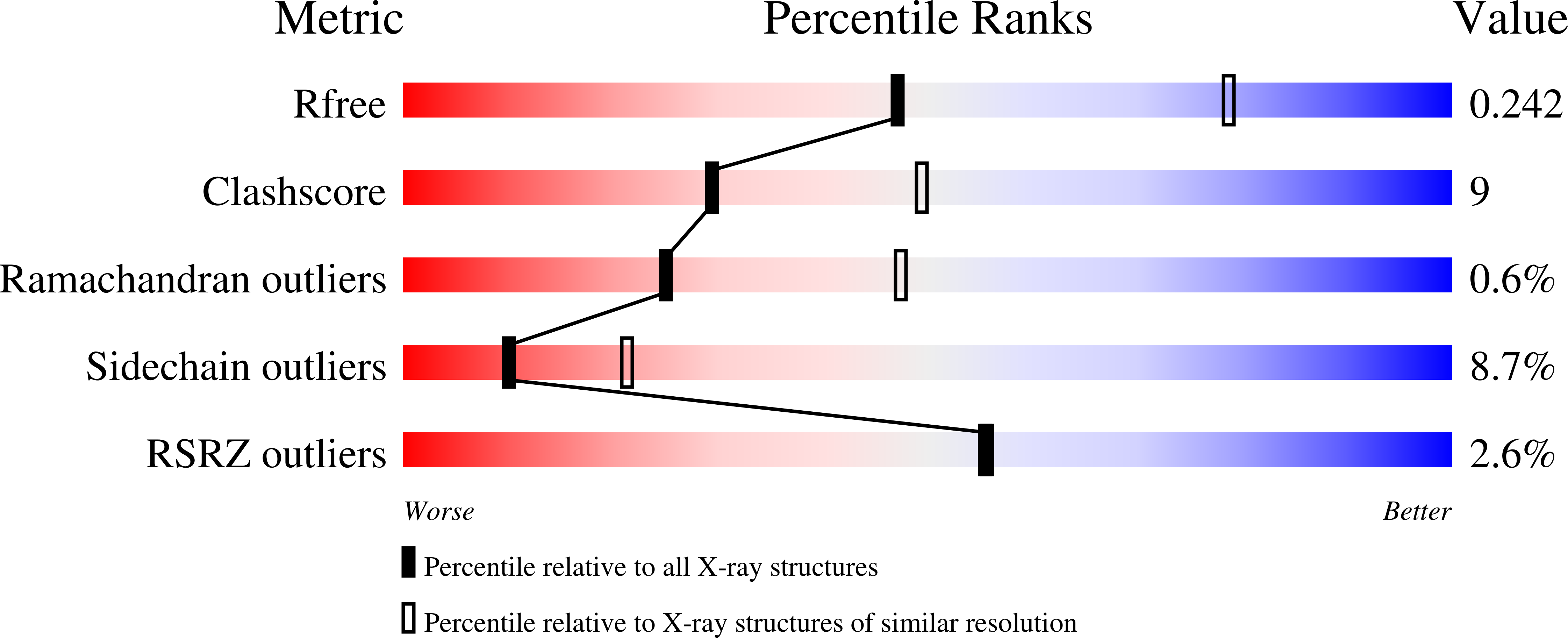
Phosphodeoxyribosyltransferases, designed enzymes for deoxyribonucleotides synthesis.
Kaminski, P.A., Labesse, G.(2013) J Biol Chem 288: 6534-6541
- PubMed: 23325804
- DOI: https://doi.org/10.1074/jbc.M112.446492
- Primary Citation of Related Structures:
4HX9 - PubMed Abstract:
A large number of nucleoside analogues and 2'-deoxynucleoside triphosphates (dNTP) have been synthesized to interfere with DNA metabolism. However, in vivo the concentration and phosphorylation of these analogues are key limiting factors. In this context, we designed enzymes to switch nucleobases attached to a deoxyribose monophosphate. Active chimeras were made from two distantly related enzymes: a nucleoside deoxyribosyltransferase from lactobacilli and a 5'-monophosphate-2'-deoxyribonucleoside hydrolase from rat. Then their unprecedented activity was further extended to deoxyribose triphosphate, and in vitro biosyntheses could be successfully performed with several base analogues. These new enzymes provide new tools to synthesize dNTP analogues and to deliver them into cells.
Organizational Affiliation:
Institut Pasteur, Unité de Chimie et Biocatalyse, CNRS, UMR 3523, 75724 Paris cedex 15, France. pierre-alexandre.kaminski@pasteur.fr