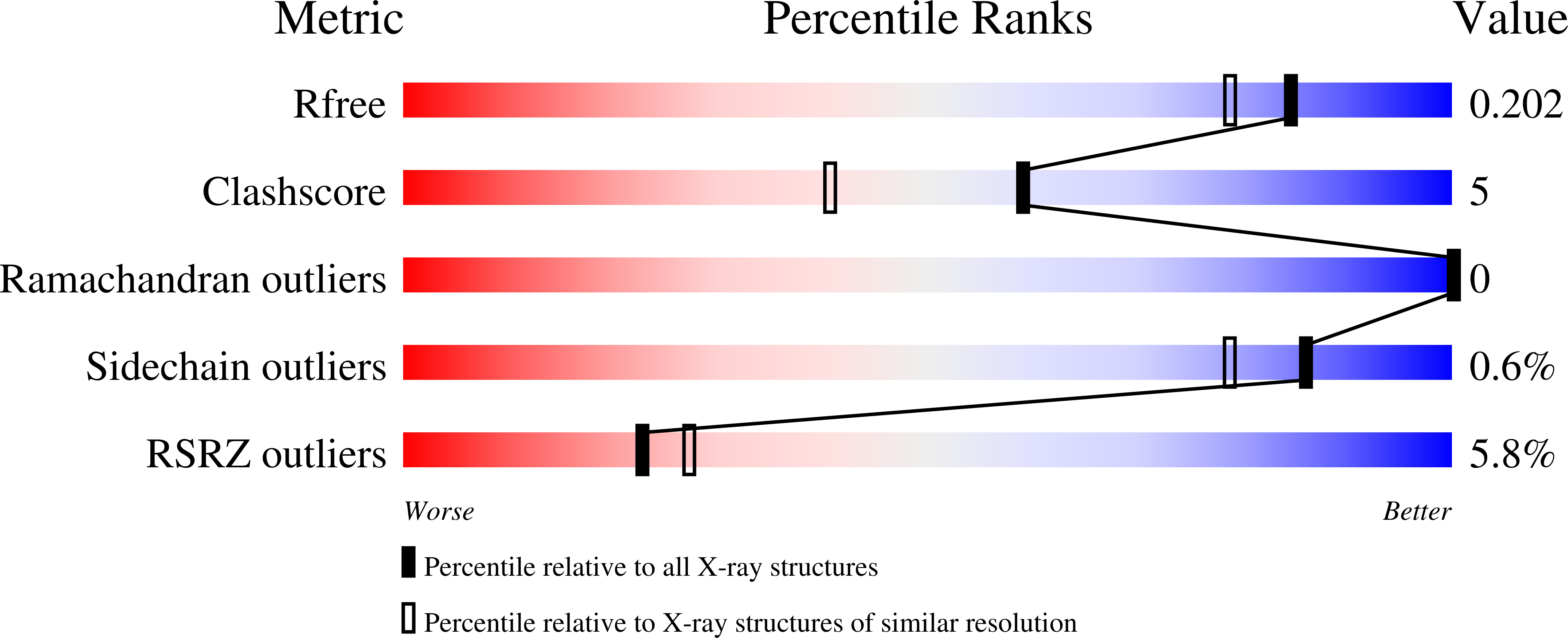
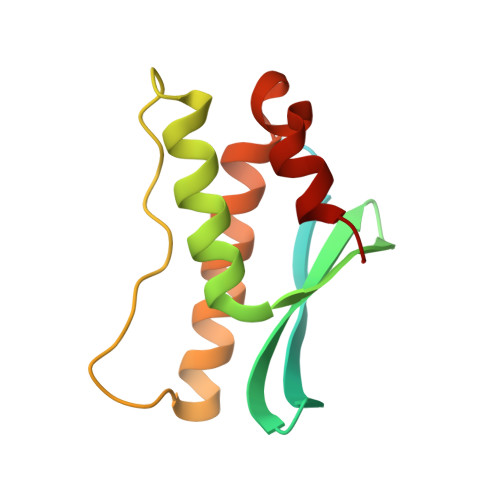
Crystal structure of PX domain of human sorting nexin SNX27
Froese, D.S., Krojer, T., Strain-Damerell, C., Allerston, C., Kiyani, W., Burgess-Brown, N., von Delft, F., Arrowsmith, C., Bountra, C., Edwards, A., Yue, W.W., Structural Genomics Consortium (SGC)To be published.