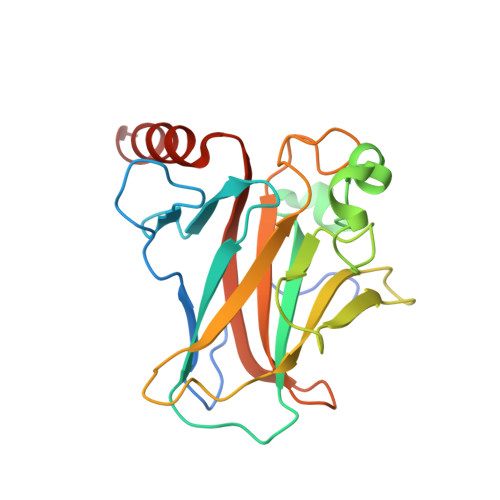
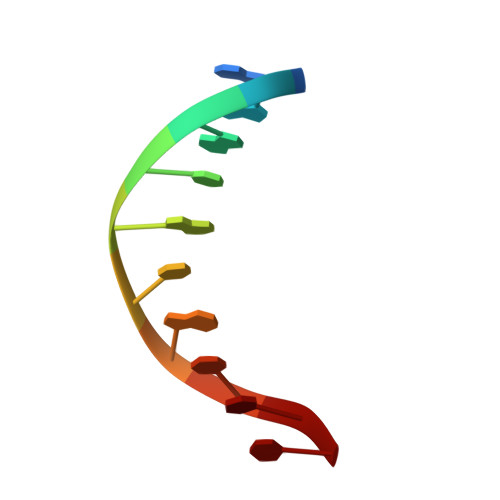
Transactivation specificity is conserved among p53 family proteins and depends on a response element sequence code.
Ciribilli, Y., Monti, P., Bisio, A., Nguyen, H.T., Ethayathulla, A.S., Ramos, A., Foggetti, G., Menichini, P., Menendez, D., Resnick, M.A., Viadiu, H., Fronza, G., Inga, A.(2013) Nucleic Acids Res 41: 8637-8653
- PubMed: 23892287
- DOI: https://doi.org/10.1093/nar/gkt657
- Primary Citation of Related Structures:
4GUQ - PubMed Abstract:
Structural and biochemical studies have demonstrated that p73, p63 and p53 recognize DNA with identical amino acids and similar binding affinity. Here, measuring transactivation activity for a large number of response elements (REs) in yeast and human cell lines, we show that p53 family proteins also have overlapping transactivation profiles. We identified mutations at conserved amino acids of loops L1 and L3 in the DNA-binding domain that tune the transactivation potential nearly equally in p73, p63 and p53. For example, the mutant S139F in p73 has higher transactivation potential towards selected REs, enhanced DNA-binding cooperativity in vitro and a flexible loop L1 as seen in the crystal structure of the protein-DNA complex. By studying, how variations in the RE sequence affect transactivation specificity, we discovered a RE-transactivation code that predicts enhanced transactivation; this correlation is stronger for promoters of genes associated with apoptosis.
Organizational Affiliation:
Laboratory of Transcriptional Networks, Centre for Integrative Biology (CIBIO), University of Trento, TN, 38060 Italy, Molecular Mutagenesis and DNA Repair Unit, IRCSS Azienda Ospedaliera Universitaria San Martino-IST-Istituto Nazionale per la Ricerca sul Cancro, Genoa 16132, Italy, Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA, 92093, USA and Chromosome Stability Group, Laboratory of Molecular Genetics, National Institute of Environmental Health Sciences, NIEHS, NIH, RTP, NC, 27709, USA.