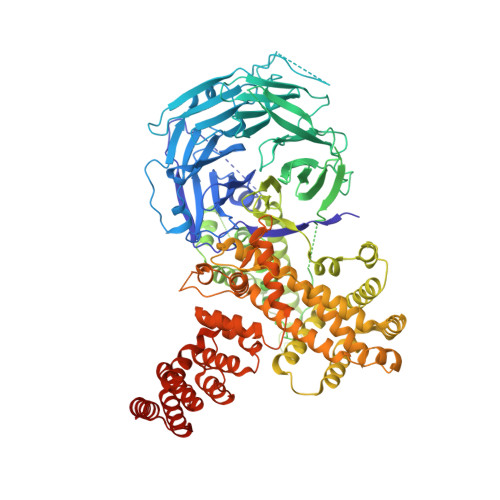
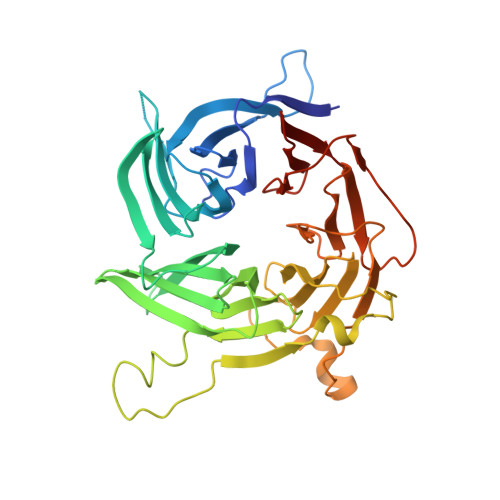
Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu, X., Mitchell, J.M., Wozniak, R.W., Blobel, G., Fan, J.(2012) Proc Natl Acad Sci U S A 109: 16498-16503
- PubMed: 23019579
- DOI: https://doi.org/10.1073/pnas.1214557109
- Primary Citation of Related Structures:
4GQ1, 4GQ2 - PubMed Abstract:
The coatomer module of the nuclear pore complex borders the cylinder-like nuclear pore-membrane domain of the nuclear envelope. In evolution, a single coatomer module increases in size from hetero-heptamer (Saccharomyces cerevisiae) to hetero-octamer (Schizosaccharomyces pombe) to hetero-nonamer (Metazoa). Notably, the heptamer-octamer transition proceeds through the acquisition of the nucleoporin Nup37. How Nup37 contacts the heptamer remained unknown. Using recombinant nucleoporins, we show that Sp-Nup37 specifically binds the Sp-Nup120 member of the hetero-heptamer but does not bind an Sc-Nup120 homolog. To elucidate the Nup37-Nup120 interaction at the atomic level, we carried out crystallographic analyses of Sp-Nup37 alone and in a complex with an N-terminal, ~110-kDa fragment of Sp-Nup120 comprising residues 1-950. Corroborating structural predictions, we determined that Nup37 folds into a seven-bladed β-propeller. Several disordered surface regions of the Nup37 β-propeller assume structure when bound to Sp-Nup120. The N-terminal domain of Sp-Nup120(1-950) also folds into a seven-bladed propeller with a markedly protruding 6D-7A insert and is followed by a contorted helical domain. Conspicuously, this 6D-7A insert contains an extension of 50 residues which also is highly conserved in Metazoa but is absent in Sc-Nup120. Strikingly, numerous contacts with the Nup37 β-propeller are located on this extension of the 6D-7A insert. Another contact region is situated toward the end of the helical region of Sp-Nup120(1-950). Our findings provide information about the evolution and the assembly of the coatomer module of the nuclear pore complex.
Organizational Affiliation:
Laboratory of Cell Biology, Rockefeller University, New York, NY 10065, USA.