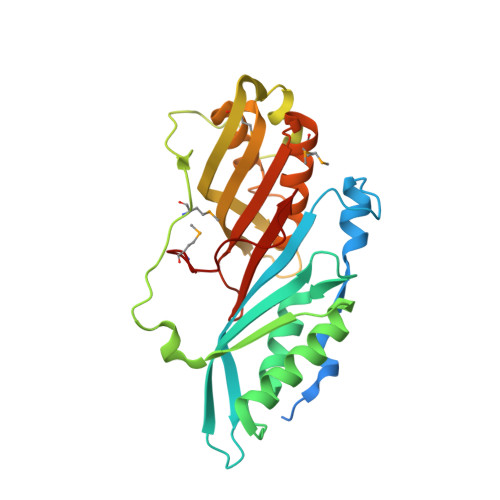
Crystal Structure of the H233A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ0
Kim, Y., Zhou, M., Gu, M., Anderson, W.F., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.